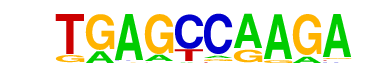
| p-value: | 1e-161 |
| log p-value: | -3.722e+02 |
| Information Content per bp: | 1.740 |
| Number of Target Sequences with motif | 19511.0 |
| Percentage of Target Sequences with motif | 24.57% |
| Number of Background Sequences with motif | 16370.8 |
| Percentage of Background Sequences with motif | 20.59% |
| Average Position of motif in Targets | 164.5 +/- 92.9bp |
| Average Position of motif in Background | 162.6 +/- 105.3bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.03 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
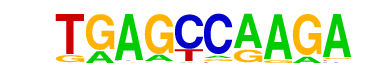
MA0099.2_JUN::FOS/Jaspar
| Match Rank: | 1 |
| Score: | 0.64
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TGAGCCAAGA
TGAGTCA--- |
|

|
|
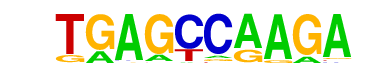
MA0161.1_NFIC/Jaspar
| Match Rank: | 2 |
| Score: | 0.61
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | TGAGCCAAGA
--TGCCAA-- |
|

|
|
AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 3 |
| Score: | 0.59
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --TGAGCCAAGA
GATGAGTCAT-- |
|


|
|
Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer
| Match Rank: | 4 |
| Score: | 0.59
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---TGAGCCAAGA
TGCTGAGTCA--- |
|


|
|
BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer
| Match Rank: | 5 |
| Score: | 0.58
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --TGAGCCAAGA
DATGASTCAT-- |
|


|
|
MA0462.1_BATF::JUN/Jaspar
| Match Rank: | 6 |
| Score: | 0.58
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----TGAGCCAAGA
GAAATGACTCA--- |
|


|
|
SD0002.1_at_AC_acceptor/Jaspar
| Match Rank: | 7 |
| Score: | 0.58
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | TGAGCCAAGA--
-AAGGCAAGTGT |
|


|
|
MA0029.1_Mecom/Jaspar
| Match Rank: | 8 |
| Score: | 0.57
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | TGAGCCAAGA-----
-AAGATAAGATAACA |
|


|
|
Fox:Ebox(Forkhead,bHLH)/Panc1-Foxa2-ChIP-Seq(GSE47459)/Homer
| Match Rank: | 9 |
| Score: | 0.56
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----TGAGCCAAGA--
NNNVCTGWGYAAACASN |
|


|
|
MA0477.1_FOSL1/Jaspar
| Match Rank: | 10 |
| Score: | 0.56
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --TGAGCCAAGA
GGTGACTCATG- |
|


|
|