| p-value: | 1e-195 |
| log p-value: | -4.492e+02 |
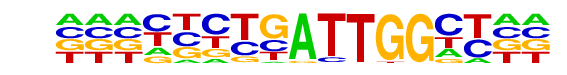
| Information Content per bp: | 1.616 |
| Number of Target Sequences with motif | 3663.0 |
| Percentage of Target Sequences with motif | 10.61% |
| Number of Background Sequences with motif | 2144.4 |
| Percentage of Background Sequences with motif | 6.35% |
| Average Position of motif in Targets | 299.2 +/- 119.0bp |
| Average Position of motif in Background | 257.9 +/- 152.9bp |
| Strand Bias (log2 ratio + to - strand density) | -0.0 |
| Multiplicity (# of sites on avg that occur together) | 1.27 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
NFY(CCAAT)/Promoter/Homer
| Match Rank: | 1 |
| Score: | 0.96
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | YBCTGATTGGCY
--CCGATTGGCT |
|


|
|
POL004.1_CCAAT-box/Jaspar
| Match Rank: | 2 |
| Score: | 0.94
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | YBCTGATTGGCY---
---TGATTGGCTANN |
|


|
|
MA0502.1_NFYB/Jaspar
| Match Rank: | 3 |
| Score: | 0.93
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | YBCTGATTGGCY-----
--CTGATTGGTCNATTT |
|


|
|
MA0060.2_NFYA/Jaspar
| Match Rank: | 4 |
| Score: | 0.87
| | Offset: | -4
| | Orientation: | forward strand |
| Alignment: | ----YBCTGATTGGCY--
AGAGTGCTGATTGGTCCA |
|


|
|
MA0161.1_NFIC/Jaspar
| Match Rank: | 5 |
| Score: | 0.78
| | Offset: | 6
| | Orientation: | forward strand |
| Alignment: | YBCTGATTGGCY
------TTGGCA |
|


|
|
PH0026.1_Duxbl/Jaspar
| Match Rank: | 6 |
| Score: | 0.75
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---YBCTGATTGGCY--
NNNNGTTGATTGGGTCG |
|

|
|
MA0038.1_Gfi1/Jaspar
| Match Rank: | 7 |
| Score: | 0.69
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | YBCTGATTGGCY
CNGTGATTTN-- |
|


|
|
PH0005.1_Barhl1/Jaspar
| Match Rank: | 8 |
| Score: | 0.69
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -YBCTGATTGGCY---
GNNTTAATTGGTTGTT |
|


|
|
PH0057.1_Hoxb13/Jaspar
| Match Rank: | 9 |
| Score: | 0.68
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---YBCTGATTGGCY-
NNAATTTTATTGGNTN |
|


|
|
PH0089.1_Isx/Jaspar
| Match Rank: | 10 |
| Score: | 0.68
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --YBCTGATTGGCY--
ACTCCTAATTAGTCGT |
|


|
|





















