| p-value: | 1e-16 |
| log p-value: | -3.696e+01 |
| Information Content per bp: | 1.936 |
| Number of Target Sequences with motif | 318.0 |
| Percentage of Target Sequences with motif | 1.14% |
| Number of Background Sequences with motif | 190.5 |
| Percentage of Background Sequences with motif | 0.69% |
| Average Position of motif in Targets | 107.6 +/- 59.8bp |
| Average Position of motif in Background | 119.9 +/- 63.5bp |
| Strand Bias (log2 ratio + to - strand density) | 0.1 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
MA0078.1_Sox17/Jaspar
| Match Rank: | 1 |
| Score: | 0.82
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---ATTGTCTG
CTCATTGTC-- |
|


|
|
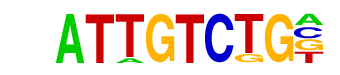
Smad3(MAD)/NPC-Smad3-ChIP-Seq(GSE36673)/Homer
| Match Rank: | 2 |
| Score: | 0.80
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | ATTGTCTG-
-TWGTCTGV |
|

|
|
PB0168.1_Sox14_2/Jaspar
| Match Rank: | 3 |
| Score: | 0.79
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----ATTGTCTG--
NNNCCATTGTGTNAN |
|


|
|
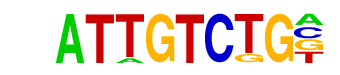
Smad2(MAD)/ES-SMAD2-ChIP-Seq(GSE29422)/Homer
| Match Rank: | 4 |
| Score: | 0.76
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | ATTGTCTG-
-CTGTCTGG |
|

|
|
PB0060.1_Smad3_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.74
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---ATTGTCTG------
NNTNNTGTCTGGNNTNG |
|


|
|
Smad4(MAD)/ESC-SMAD4-ChIP-Seq(GSE29422)/Homer
| Match Rank: | 6 |
| Score: | 0.73
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -ATTGTCTG-
VBSYGTCTGG |
|


|
|
Oct4:Sox17(POU,Homeobox,HMG)/F9-Sox17-ChIP-Seq(GSE44553)/Homer
| Match Rank: | 7 |
| Score: | 0.72
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --ATTGTCTG-----
CCATTGTATGCAAAT |
|


|
|
PB0165.1_Sox11_2/Jaspar
| Match Rank: | 8 |
| Score: | 0.71
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---ATTGTCTG---
AAAATTGTTATGAA |
|


|
|
Sox6(HMG)/Myotubes-Sox6-ChIP-Seq(GSE32627)/Homer
| Match Rank: | 9 |
| Score: | 0.71
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --ATTGTCTG
CCATTGTTNY |
|


|
|
MF0011.1_HMG_class/Jaspar
| Match Rank: | 10 |
| Score: | 0.71
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | ATTGTCTG
ATTGTT-- |
|


|
|





















