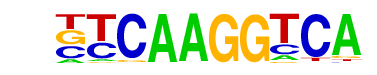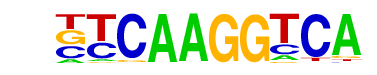| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | Sox3(HMG)/NPC-Sox3-ChIP-Seq(GSE33059)/Homer | 1e-55 | -1.274e+02 | 0.0000 | 4783.0 | 17.12% | 3789.0 | 13.78% | motif file (matrix) |
pdf |
| 2 |  | Sox10(HMG)/SciaticNerve-Sox3-ChIP-Seq(GSE35132)/Homer | 1e-51 | -1.189e+02 | 0.0000 | 4498.0 | 16.10% | 3562.8 | 12.96% | motif file (matrix) |
pdf |
| 3 |  | AR-halfsite(NR)/LNCaP-AR-ChIP-Seq(GSE27824)/Homer | 1e-43 | -1.006e+02 | 0.0000 | 12047.0 | 43.13% | 10735.4 | 39.04% | motif file (matrix) |
pdf |
| 4 | 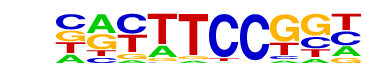 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-43 | -9.920e+01 | 0.0000 | 4411.0 | 15.79% | 3556.4 | 12.93% | motif file (matrix) |
pdf |
| 5 |  | ZFX(Zf)/mES-Zfx-ChIP-Seq(GSE11431)/Homer | 1e-41 | -9.517e+01 | 0.0000 | 5842.0 | 20.91% | 4881.4 | 17.75% | motif file (matrix) |
pdf |
| 6 |  | Sox2(HMG)/mES-Sox2-ChIP-Seq(GSE11431)/Homer | 1e-36 | -8.417e+01 | 0.0000 | 2434.0 | 8.71% | 1850.2 | 6.73% | motif file (matrix) |
pdf |
| 7 |  | Sox4(HMG)/proB-Sox4-ChIP-Seq(GSE50066)/Homer | 1e-32 | -7.418e+01 | 0.0000 | 2369.0 | 8.48% | 1825.7 | 6.64% | motif file (matrix) |
pdf |
| 8 | 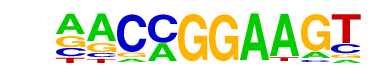 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-29 | -6.765e+01 | 0.0000 | 5018.0 | 17.96% | 4251.4 | 15.46% | motif file (matrix) |
pdf |
| 9 |  | EBF1(EBF)/Near-E2A-ChIP-Seq(GSE21512)/Homer | 1e-29 | -6.753e+01 | 0.0000 | 4709.0 | 16.86% | 3966.5 | 14.42% | motif file (matrix) |
pdf |
| 10 |  | ZNF711(Zf)/SHSY5Y-ZNF711-ChIP-Seq(GSE20673)/Homer | 1e-28 | -6.642e+01 | 0.0000 | 8593.0 | 30.76% | 7623.3 | 27.72% | motif file (matrix) |
pdf |
| 11 |  | Fox:Ebox(Forkhead,bHLH)/Panc1-Foxa2-ChIP-Seq(GSE47459)/Homer | 1e-28 | -6.557e+01 | 0.0000 | 2468.0 | 8.84% | 1942.1 | 7.06% | motif file (matrix) |
pdf |
| 12 |  | MYB(HTH)/ERMYB-Myb-ChIPSeq(GSE22095)/Homer | 1e-27 | -6.313e+01 | 0.0000 | 5151.0 | 18.44% | 4398.2 | 15.99% | motif file (matrix) |
pdf |
| 13 |  | AP-2gamma(AP2)/MCF7-TFAP2C-ChIP-Seq(GSE21234)/Homer | 1e-26 | -6.054e+01 | 0.0000 | 5137.0 | 18.39% | 4399.7 | 16.00% | motif file (matrix) |
pdf |
| 14 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-26 | -6.031e+01 | 0.0000 | 3443.0 | 12.33% | 2840.0 | 10.33% | motif file (matrix) |
pdf |
| 15 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-25 | -5.937e+01 | 0.0000 | 5440.0 | 19.48% | 4688.6 | 17.05% | motif file (matrix) |
pdf |
| 16 |  | BMYB(HTH)/Hela-BMYB-ChIP-Seq(GSE27030)/Homer | 1e-25 | -5.804e+01 | 0.0000 | 4154.0 | 14.87% | 3503.0 | 12.74% | motif file (matrix) |
pdf |
| 17 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-25 | -5.772e+01 | 0.0000 | 2586.0 | 9.26% | 2077.7 | 7.56% | motif file (matrix) |
pdf |
| 18 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-24 | -5.684e+01 | 0.0000 | 2630.0 | 9.42% | 2120.0 | 7.71% | motif file (matrix) |
pdf |
| 19 |  | AP-2alpha(AP2)/Hela-AP2alpha-ChIP-Seq(GSE31477)/Homer | 1e-22 | -5.228e+01 | 0.0000 | 3985.0 | 14.27% | 3376.2 | 12.28% | motif file (matrix) |
pdf |
| 20 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-21 | -5.050e+01 | 0.0000 | 2406.0 | 8.61% | 1945.0 | 7.07% | motif file (matrix) |
pdf |
| 21 |  | AMYB(HTH)/Testes-AMYB-ChIP-Seq(GSE44588)/Homer | 1e-21 | -4.859e+01 | 0.0000 | 4270.0 | 15.29% | 3660.6 | 13.31% | motif file (matrix) |
pdf |
| 22 |  | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-20 | -4.758e+01 | 0.0000 | 4211.0 | 15.08% | 3611.7 | 13.13% | motif file (matrix) |
pdf |
| 23 |  | Sox6(HMG)/Myotubes-Sox6-ChIP-Seq(GSE32627)/Homer | 1e-20 | -4.707e+01 | 0.0000 | 4030.0 | 14.43% | 3446.3 | 12.53% | motif file (matrix) |
pdf |
| 24 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-19 | -4.516e+01 | 0.0000 | 3757.0 | 13.45% | 3204.1 | 11.65% | motif file (matrix) |
pdf |
| 25 |  | Isl1(Homeobox)/Neuron-Isl1-ChIP-Seq(GSE31456)/Homer | 1e-16 | -3.835e+01 | 0.0000 | 4767.0 | 17.07% | 4188.3 | 15.23% | motif file (matrix) |
pdf |
| 26 |  | IRF2(IRF)/Erythroblas-IRF2-ChIP-Seq(GSE36985)/Homer | 1e-16 | -3.774e+01 | 0.0000 | 360.0 | 1.29% | 221.0 | 0.80% | motif file (matrix) |
pdf |
| 27 |  | Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-15 | -3.610e+01 | 0.0000 | 931.0 | 3.33% | 696.6 | 2.53% | motif file (matrix) |
pdf |
| 28 |  | ISRE(IRF)/ThioMac-LPS-Expression(GSE23622)/Homer | 1e-14 | -3.421e+01 | 0.0000 | 230.0 | 0.82% | 128.0 | 0.47% | motif file (matrix) |
pdf |
| 29 |  | EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer | 1e-13 | -3.222e+01 | 0.0000 | 2061.0 | 7.38% | 1716.9 | 6.24% | motif file (matrix) |
pdf |
| 30 | 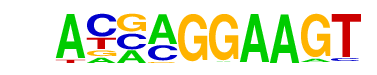 | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-13 | -3.172e+01 | 0.0000 | 2416.0 | 8.65% | 2043.5 | 7.43% | motif file (matrix) |
pdf |
| 31 |  | PR(NR)/T47D-PR-ChIP-Seq(GSE31130)/Homer | 1e-13 | -3.060e+01 | 0.0000 | 5987.0 | 21.43% | 5402.4 | 19.65% | motif file (matrix) |
pdf |
| 32 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-12 | -2.908e+01 | 0.0000 | 1031.0 | 3.69% | 806.4 | 2.93% | motif file (matrix) |
pdf |
| 33 |  | Nr5a2(NR)/mES-Nr5a2-ChIP-Seq(GSE19019)/Homer | 1e-11 | -2.754e+01 | 0.0000 | 1596.0 | 5.71% | 1317.5 | 4.79% | motif file (matrix) |
pdf |
| 34 |  | PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-11 | -2.743e+01 | 0.0000 | 1591.0 | 5.70% | 1313.9 | 4.78% | motif file (matrix) |
pdf |
| 35 | 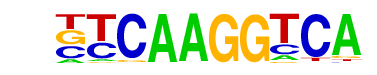 | Nr5a2(NR)/Pancreas-LRH1-ChIP-Seq(GSE34295)/Homer | 1e-11 | -2.737e+01 | 0.0000 | 2113.0 | 7.56% | 1789.3 | 6.51% | motif file (matrix) |
pdf |
| 36 |  | n-Myc(bHLH)/mES-nMyc-ChIP-Seq(GSE11431)/Homer | 1e-11 | -2.602e+01 | 0.0000 | 2375.0 | 8.50% | 2038.3 | 7.41% | motif file (matrix) |
pdf |
| 37 |  | Nrf2(bZIP)/Lymphoblast-Nrf2-ChIP-Seq(GSE37589)/Homer | 1e-10 | -2.527e+01 | 0.0000 | 132.0 | 0.47% | 68.1 | 0.25% | motif file (matrix) |
pdf |
| 38 | 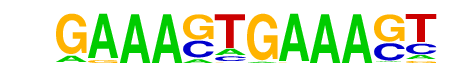 | IRF1(IRF)/PBMC-IRF1-ChIP-Seq(GSE43036)/Homer | 1e-10 | -2.519e+01 | 0.0000 | 427.0 | 1.53% | 299.8 | 1.09% | motif file (matrix) |
pdf |
| 39 |  | ETS(ETS)/Promoter/Homer | 1e-10 | -2.504e+01 | 0.0000 | 1449.0 | 5.19% | 1196.9 | 4.35% | motif file (matrix) |
pdf |
| 40 |  | NF-E2(bZIP)/K562-NFE2-ChIP-Seq(GSE31477)/Homer | 1e-10 | -2.410e+01 | 0.0000 | 147.0 | 0.53% | 80.1 | 0.29% | motif file (matrix) |
pdf |
| 41 |  | RXR(NR),DR1/3T3L1-RXR-ChIP-Seq(GSE13511)/Homer | 1e-10 | -2.369e+01 | 0.0000 | 3749.0 | 13.42% | 3338.6 | 12.14% | motif file (matrix) |
pdf |
| 42 |  | Foxa2(Forkhead)/Liver-Foxa2-ChIP-Seq(GSE25694)/Homer | 1e-9 | -2.246e+01 | 0.0000 | 1806.0 | 6.47% | 1535.3 | 5.58% | motif file (matrix) |
pdf |
| 43 |  | BMAL1(bHLH)/Liver-Bmal1-ChIP-Seq(GSE39860)/Homer | 1e-9 | -2.153e+01 | 0.0000 | 5206.0 | 18.64% | 4740.1 | 17.24% | motif file (matrix) |
pdf |
| 44 |  | Esrrb(NR)/mES-Esrrb-ChIP-Seq(GSE11431)/Homer | 1e-9 | -2.124e+01 | 0.0000 | 1909.0 | 6.83% | 1637.7 | 5.96% | motif file (matrix) |
pdf |
| 45 |  | Max(bHLH)/K562-Max-ChIP-Seq(GSE31477)/Homer | 1e-8 | -2.011e+01 | 0.0000 | 2084.0 | 7.46% | 1806.5 | 6.57% | motif file (matrix) |
pdf |
| 46 |  | Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan et al.)/Homer | 1e-8 | -1.986e+01 | 0.0000 | 4975.0 | 17.81% | 4537.0 | 16.50% | motif file (matrix) |
pdf |
| 47 |  | Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer | 1e-8 | -1.919e+01 | 0.0000 | 478.0 | 1.71% | 358.6 | 1.30% | motif file (matrix) |
pdf |
| 48 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-8 | -1.903e+01 | 0.0000 | 1867.0 | 6.68% | 1612.9 | 5.87% | motif file (matrix) |
pdf |
| 49 |  | c-Myc(bHLH)/mES-cMyc-ChIP-Seq(GSE11431)/Homer | 1e-7 | -1.834e+01 | 0.0000 | 1636.0 | 5.86% | 1403.5 | 5.10% | motif file (matrix) |
pdf |
| 50 |  | SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer | 1e-7 | -1.767e+01 | 0.0000 | 3207.0 | 11.48% | 2878.0 | 10.47% | motif file (matrix) |
pdf |
| 51 |  | EBF(EBF)/proBcell-EBF-ChIP-Seq(GSE21978)/Homer | 1e-7 | -1.734e+01 | 0.0000 | 891.0 | 3.19% | 729.1 | 2.65% | motif file (matrix) |
pdf |
| 52 |  | Fosl2(bZIP)/3T3L1-Fosl2-ChIP-Seq(GSE56872)/Homer | 1e-7 | -1.734e+01 | 0.0000 | 795.0 | 2.85% | 643.4 | 2.34% | motif file (matrix) |
pdf |
| 53 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-7 | -1.638e+01 | 0.0000 | 1523.0 | 5.45% | 1311.0 | 4.77% | motif file (matrix) |
pdf |
| 54 |  | Fra1(bZIP)/BT549-Fra1-ChIP-Seq(GSE46166)/Homer | 1e-6 | -1.497e+01 | 0.0000 | 1277.0 | 4.57% | 1093.9 | 3.98% | motif file (matrix) |
pdf |
| 55 |  | PPARE(NR),DR1/3T3L1-Pparg-ChIP-Seq(GSE13511)/Homer | 1e-6 | -1.445e+01 | 0.0000 | 2992.0 | 10.71% | 2703.4 | 9.83% | motif file (matrix) |
pdf |
| 56 |  | HNF4a(NR),DR1/HepG2-HNF4a-ChIP-Seq(GSE25021)/Homer | 1e-6 | -1.429e+01 | 0.0000 | 1216.0 | 4.35% | 1042.0 | 3.79% | motif file (matrix) |
pdf |
| 57 |  | USF1(bHLH)/GM12878-Usf1-ChIP-Seq(GSE32465)/Homer | 1e-6 | -1.393e+01 | 0.0000 | 1484.0 | 5.31% | 1291.7 | 4.70% | motif file (matrix) |
pdf |
| 58 |  | STAT4(Stat)/CD4-Stat4-ChIP-Seq(GSE22104)/Homer | 1e-5 | -1.331e+01 | 0.0000 | 2310.0 | 8.27% | 2069.4 | 7.52% | motif file (matrix) |
pdf |
| 59 |  | YY1(Zf)/Promoter/Homer | 1e-5 | -1.325e+01 | 0.0000 | 366.0 | 1.31% | 280.7 | 1.02% | motif file (matrix) |
pdf |
| 60 |  | Smad3(MAD)/NPC-Smad3-ChIP-Seq(GSE36673)/Homer | 1e-5 | -1.304e+01 | 0.0000 | 8337.0 | 29.85% | 7865.0 | 28.60% | motif file (matrix) |
pdf |
| 61 |  | RORgt(NR)/EL4-RORgt.Flag-ChIP-Seq(GSE56019)/Homer | 1e-5 | -1.292e+01 | 0.0000 | 301.0 | 1.08% | 225.9 | 0.82% | motif file (matrix) |
pdf |
| 62 |  | Oct4:Sox17(POU,Homeobox,HMG)/F9-Sox17-ChIP-Seq(GSE44553)/Homer | 1e-5 | -1.278e+01 | 0.0000 | 311.0 | 1.11% | 234.4 | 0.85% | motif file (matrix) |
pdf |
| 63 |  | TR4(NR),DR1/Hela-TR4-ChIP-Seq(GSE24685)/Homer | 1e-5 | -1.270e+01 | 0.0000 | 342.0 | 1.22% | 261.7 | 0.95% | motif file (matrix) |
pdf |
| 64 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-5 | -1.258e+01 | 0.0000 | 1702.0 | 6.09% | 1504.5 | 5.47% | motif file (matrix) |
pdf |
| 65 |  | Nkx6.1(Homeobox)/Islet-Nkx6.1-ChIP-Seq(GSE40975)/Homer | 1e-5 | -1.227e+01 | 0.0000 | 5704.0 | 20.42% | 5325.8 | 19.37% | motif file (matrix) |
pdf |
| 66 |  | Tcf4(HMG)/Hct116-Tcf4-ChIP-Seq(SRA012054)/Homer | 1e-5 | -1.195e+01 | 0.0000 | 958.0 | 3.43% | 818.2 | 2.98% | motif file (matrix) |
pdf |
| 67 |  | Stat3(Stat)/mES-Stat3-ChIP-Seq(GSE11431)/Homer | 1e-5 | -1.182e+01 | 0.0000 | 1502.0 | 5.38% | 1323.5 | 4.81% | motif file (matrix) |
pdf |
| 68 |  | MITF(bHLH)/MastCells-MITF-ChIP-Seq(GSE48085)/Homer | 1e-5 | -1.178e+01 | 0.0000 | 2650.0 | 9.49% | 2405.8 | 8.75% | motif file (matrix) |
pdf |
| 69 |  | Foxh1(Forkhead)/hESC-FOXH1-ChIP-Seq(GSE29422)/Homer | 1e-5 | -1.169e+01 | 0.0000 | 1427.0 | 5.11% | 1254.8 | 4.56% | motif file (matrix) |
pdf |
| 70 |  | c-Myc(bHLH)/LNCAP-cMyc-ChIP-Seq(Unpublished)/Homer | 1e-5 | -1.159e+01 | 0.0000 | 1782.0 | 6.38% | 1587.3 | 5.77% | motif file (matrix) |
pdf |
| 71 |  | SpiB(ETS)/OCILY3-SPIB-ChIP-Seq(GSE56857)/Homer | 1e-4 | -1.133e+01 | 0.0000 | 752.0 | 2.69% | 633.4 | 2.30% | motif file (matrix) |
pdf |
| 72 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-4 | -1.094e+01 | 0.0001 | 580.0 | 2.08% | 479.5 | 1.74% | motif file (matrix) |
pdf |
| 73 |  | VDR(NR),DR3/GM10855-VDR+vitD-ChIP-Seq(GSE22484)/Homer | 1e-4 | -1.091e+01 | 0.0001 | 655.0 | 2.34% | 547.3 | 1.99% | motif file (matrix) |
pdf |
| 74 |  | Smad2(MAD)/ES-SMAD2-ChIP-Seq(GSE29422)/Homer | 1e-4 | -1.062e+01 | 0.0001 | 5062.0 | 18.12% | 4730.0 | 17.20% | motif file (matrix) |
pdf |
| 75 |  | RUNX(Runt)/HPC7-Runx1-ChIP-Seq(GSE22178)/Homer | 1e-4 | -9.942e+00 | 0.0002 | 1862.0 | 6.67% | 1677.5 | 6.10% | motif file (matrix) |
pdf |
| 76 |  | Stat3+il21(Stat)/CD4-Stat3-ChIP-Seq(GSE19198)/Homer | 1e-4 | -9.907e+00 | 0.0002 | 1868.0 | 6.69% | 1683.2 | 6.12% | motif file (matrix) |
pdf |
| 77 |  | Bach1(bZIP)/K562-Bach1-ChIP-Seq(GSE31477)/Homer | 1e-4 | -9.706e+00 | 0.0002 | 121.0 | 0.43% | 82.8 | 0.30% | motif file (matrix) |
pdf |
| 78 |  | STAT5(Stat)/mCD4+-Stat5-ChIP-Seq(GSE12346)/Homer | 1e-4 | -9.658e+00 | 0.0002 | 783.0 | 2.80% | 671.5 | 2.44% | motif file (matrix) |
pdf |
| 79 |  | Bcl6(Zf)/Liver-Bcl6-ChIP-Seq(GSE31578)/Homer | 1e-4 | -9.533e+00 | 0.0002 | 3448.0 | 12.34% | 3192.2 | 11.61% | motif file (matrix) |
pdf |
| 80 |  | NFkB-p65(RHD)/GM12787-p65-ChIP-Seq(GSE19485)/Homer | 1e-3 | -9.191e+00 | 0.0003 | 1562.0 | 5.59% | 1401.8 | 5.10% | motif file (matrix) |
pdf |
| 81 |  | STAT6(Stat)/Macrophage-Stat6-ChIP-Seq(GSE38377)/Homer | 1e-3 | -8.868e+00 | 0.0005 | 1197.0 | 4.29% | 1061.0 | 3.86% | motif file (matrix) |
pdf |
| 82 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-3 | -8.819e+00 | 0.0005 | 1465.0 | 5.24% | 1313.8 | 4.78% | motif file (matrix) |
pdf |
| 83 |  | RUNX2(Runt)/PCa-RUNX2-ChIP-Seq(GSE33889)/Homer | 1e-3 | -8.202e+00 | 0.0009 | 2150.0 | 7.70% | 1968.3 | 7.16% | motif file (matrix) |
pdf |
| 84 |  | E2F(E2F)/Hela-CellCycle-Expression/Homer | 1e-3 | -8.119e+00 | 0.0009 | 213.0 | 0.76% | 164.4 | 0.60% | motif file (matrix) |
pdf |
| 85 |  | IRF4(IRF)/GM12878-IRF4-ChIP-Seq(GSE32465)/Homer | 1e-3 | -8.072e+00 | 0.0010 | 1017.0 | 3.64% | 899.3 | 3.27% | motif file (matrix) |
pdf |
| 86 |  | GRE(NR),IR3/A549-GR-ChIP-Seq(GSE32465)/Homer | 1e-3 | -7.920e+00 | 0.0011 | 360.0 | 1.29% | 295.9 | 1.08% | motif file (matrix) |
pdf |
| 87 |  | HIF-1b(HLH)/T47D-HIF1b-ChIP-Seq(GSE59937)/Homer | 1e-3 | -7.862e+00 | 0.0012 | 3628.0 | 12.99% | 3388.7 | 12.32% | motif file (matrix) |
pdf |
| 88 |  | STAT6(Stat)/CD4-Stat6-ChIP-Seq(GSE22104)/Homer | 1e-3 | -7.570e+00 | 0.0015 | 1146.0 | 4.10% | 1024.5 | 3.73% | motif file (matrix) |
pdf |
| 89 |  | PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-3 | -7.562e+00 | 0.0015 | 3888.0 | 13.92% | 3643.9 | 13.25% | motif file (matrix) |
pdf |
| 90 |  | Gata4(Zf)/Heart-Gata4-ChIP-Seq(GSE35151)/Homer | 1e-3 | -7.327e+00 | 0.0019 | 2310.0 | 8.27% | 2131.8 | 7.75% | motif file (matrix) |
pdf |
| 91 |  | RUNX-AML(Runt)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-3 | -6.998e+00 | 0.0027 | 1874.0 | 6.71% | 1719.3 | 6.25% | motif file (matrix) |
pdf |
| 92 |  | bZIP:IRF(bZIP,IRF)/Th17-BatF-ChIP-Seq(GSE39756)/Homer | 1e-2 | -6.838e+00 | 0.0031 | 954.0 | 3.42% | 850.2 | 3.09% | motif file (matrix) |
pdf |
| 93 |  | CLOCK(bHLH)/Liver-Clock-ChIP-Seq(GSE39860)/Homer | 1e-2 | -6.478e+00 | 0.0044 | 1707.0 | 6.11% | 1566.1 | 5.69% | motif file (matrix) |
pdf |
| 94 |  | Gata2(Zf)/K562-GATA2-ChIP-Seq(GSE18829)/Homer | 1e-2 | -6.453e+00 | 0.0044 | 1508.0 | 5.40% | 1377.3 | 5.01% | motif file (matrix) |
pdf |
| 95 |  | p53(p53)/Saos-p53-ChIP-Seq(GSE15780)/Homer | 1e-2 | -6.266e+00 | 0.0052 | 201.0 | 0.72% | 160.5 | 0.58% | motif file (matrix) |
pdf |
| 96 |  | p53(p53)/Saos-p53-ChIP-Seq/Homer | 1e-2 | -6.266e+00 | 0.0052 | 201.0 | 0.72% | 160.5 | 0.58% | motif file (matrix) |
pdf |
| 97 |  | ARE(NR)/LNCAP-AR-ChIP-Seq(GSE27824)/Homer | 1e-2 | -6.240e+00 | 0.0053 | 644.0 | 2.31% | 565.1 | 2.05% | motif file (matrix) |
pdf |
| 98 |  | Gata1(Zf)/K562-GATA1-ChIP-Seq(GSE18829)/Homer | 1e-2 | -6.214e+00 | 0.0054 | 1342.0 | 4.80% | 1222.9 | 4.45% | motif file (matrix) |
pdf |
| 99 |  | SCL(bHLH)/HPC7-Scl-ChIP-Seq(GSE13511)/Homer | 1e-2 | -5.960e+00 | 0.0069 | 12834.0 | 45.95% | 12405.5 | 45.11% | motif file (matrix) |
pdf |
| 100 |  | TATA-Box(TBP)/Promoter/Homer | 1e-2 | -5.879e+00 | 0.0074 | 3024.0 | 10.83% | 2837.6 | 10.32% | motif file (matrix) |
pdf |
| 101 |  | HRE(HSF)/Striatum-HSF1-ChIP-Seq(GSE38000)/Homer | 1e-2 | -5.870e+00 | 0.0074 | 486.0 | 1.74% | 422.0 | 1.53% | motif file (matrix) |
pdf |
| 102 |  | GRE(NR),IR3/RAW264.7-GRE-ChIP-Seq(Unpublished)/Homer | 1e-2 | -5.762e+00 | 0.0081 | 593.0 | 2.12% | 521.5 | 1.90% | motif file (matrix) |
pdf |
| 103 |  | FOXA1(Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-2 | -5.677e+00 | 0.0088 | 2520.0 | 9.02% | 2355.2 | 8.56% | motif file (matrix) |
pdf |
| 104 |  | Erra(NR)/HepG2-Erra-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.618e+00 | 0.0092 | 7187.0 | 25.73% | 6883.1 | 25.03% | motif file (matrix) |
pdf |
| 105 |  | Znf263(Zf)/K562-Znf263-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.581e+00 | 0.0095 | 7020.0 | 25.13% | 6721.7 | 24.44% | motif file (matrix) |
pdf |
| 106 |  | HRE(HSF)/HepG2-HSF1-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.451e+00 | 0.0107 | 391.0 | 1.40% | 336.2 | 1.22% | motif file (matrix) |
pdf |
| 107 |  | GATA:SCL(Zf,bHLH)/Ter119-SCL-ChIP-Seq(GSE18720)/Homer | 1e-2 | -5.431e+00 | 0.0108 | 254.0 | 0.91% | 211.2 | 0.77% | motif file (matrix) |
pdf |
| 108 |  | Smad4(MAD)/ESC-SMAD4-ChIP-Seq(GSE29422)/Homer | 1e-2 | -5.270e+00 | 0.0126 | 5195.0 | 18.60% | 4951.8 | 18.01% | motif file (matrix) |
pdf |
| 109 |  | CHR(?)/Hela-CellCycle-Expression/Homer | 1e-2 | -5.239e+00 | 0.0128 | 1235.0 | 4.42% | 1131.9 | 4.12% | motif file (matrix) |
pdf |
| 110 |  | Six1(Homeobox)/Myoblast-Six1-ChIP-Chip(GSE20150)/Homer | 1e-2 | -5.220e+00 | 0.0130 | 534.0 | 1.91% | 470.8 | 1.71% | motif file (matrix) |
pdf |
| 111 |  | Rbpj1(?)/Panc1-Rbpj1-ChIP-Seq(GSE47459)/Homer | 1e-2 | -5.107e+00 | 0.0144 | 4309.0 | 15.43% | 4094.1 | 14.89% | motif file (matrix) |
pdf |
| 112 |  | ERE(NR),IR3/MCF7-ERa-ChIP-Seq(Unpublished)/Homer | 1e-2 | -5.042e+00 | 0.0152 | 764.0 | 2.74% | 687.5 | 2.50% | motif file (matrix) |
pdf |
| 113 |  | MafA(bZIP)/Islet-MafA-ChIP-Seq(GSE30298)/Homer | 1e-2 | -4.975e+00 | 0.0161 | 1996.0 | 7.15% | 1862.6 | 6.77% | motif file (matrix) |
pdf |
| 114 |  | RUNX1(Runt)/Jurkat-RUNX1-ChIP-Seq(GSE29180)/Homer | 1e-2 | -4.940e+00 | 0.0166 | 2538.0 | 9.09% | 2384.6 | 8.67% | motif file (matrix) |
pdf |
| 115 |  | NF1:FOXA1(CTF,Forkhead)/LNCAP-FOXA1-ChIP-Seq(GSE27824)/Homer | 1e-2 | -4.826e+00 | 0.0184 | 88.0 | 0.32% | 66.1 | 0.24% | motif file (matrix) |
pdf |
| 116 |  | NF1-halfsite(CTF)/LNCaP-NF1-ChIP-Seq(Unpublished)/Homer | 1e-2 | -4.622e+00 | 0.0224 | 4852.0 | 17.37% | 4632.9 | 16.85% | motif file (matrix) |
pdf |