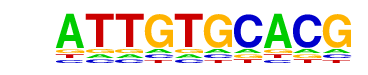
| p-value: | 1e-3 |
| log p-value: | -7.174e+00 |
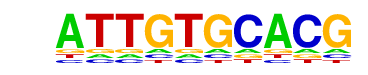
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 1.0 |
| Percentage of Target Sequences with motif | 5.56% |
| Number of Background Sequences with motif | 2.7 |
| Percentage of Background Sequences with motif | 0.01% |
| Average Position of motif in Targets | 172.0 +/- 0.0bp |
| Average Position of motif in Background | 252.3 +/- 95.6bp |
| Strand Bias (log2 ratio + to - strand density) | 10.0 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PB0168.1_Sox14_2/Jaspar
| Match Rank: | 1 |
| Score: | 0.71
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----ATTGTGCACG
NNNCCATTGTGTNAN |
|


|
|
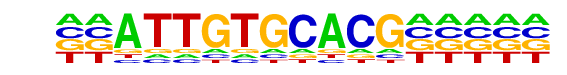
PB0104.1_Zscan4_1/Jaspar
| Match Rank: | 2 |
| Score: | 0.70
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --ATTGTGCACG-----
TACATGTGCACATAAAA |
|

|
|
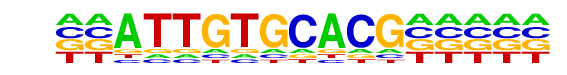
PB0026.1_Gm397_1/Jaspar
| Match Rank: | 3 |
| Score: | 0.69
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --ATTGTGCACG-----
CAGATGTGCACATACGT |
|

|
|
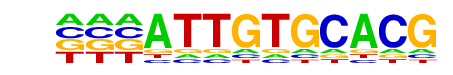
MA0078.1_Sox17/Jaspar
| Match Rank: | 4 |
| Score: | 0.68
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---ATTGTGCACG
CTCATTGTC---- |
|

|
|
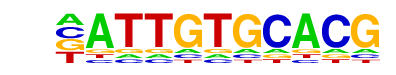
MA0102.3_CEBPA/Jaspar
| Match Rank: | 5 |
| Score: | 0.65
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -ATTGTGCACG
NATTGTGCAAT |
|

|
|
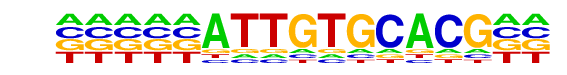
PB0173.1_Sox21_2/Jaspar
| Match Rank: | 6 |
| Score: | 0.65
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----ATTGTGCACG--
AATCAATTGTTCCGCTA |
|

|
|
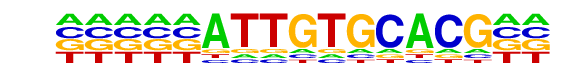
PB0183.1_Sry_2/Jaspar
| Match Rank: | 7 |
| Score: | 0.65
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----ATTGTGCACG--
CNNNTATTGTTCNNNNN |
|

|
|
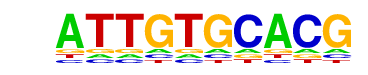
MA0084.1_SRY/Jaspar
| Match Rank: | 8 |
| Score: | 0.65
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | ATTGTGCACG
ATTGTTTAN- |
|

|
|
PB0065.1_Sox15_1/Jaspar
| Match Rank: | 9 |
| Score: | 0.65
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------ATTGTGCACG-
ANNTCTATTGTTCNNNA |
|


|
|
CEBP(bZIP)/ThioMac-CEBPb-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 10 |
| Score: | 0.65
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | ATTGTGCACG
ATTGCGCAAC |
|

|
|