| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-12 | -2.895e+01 | 0.0000 | 152.0 | 10.15% | 2312.8 | 5.43% | motif file (matrix) |
pdf |
| 2 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-9 | -2.248e+01 | 0.0000 | 143.0 | 9.55% | 2332.3 | 5.47% | motif file (matrix) |
pdf |
| 3 | 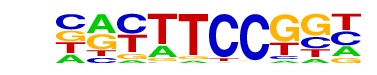 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-7 | -1.803e+01 | 0.0000 | 228.0 | 15.22% | 4494.2 | 10.54% | motif file (matrix) |
pdf |
| 4 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-6 | -1.458e+01 | 0.0000 | 119.0 | 7.94% | 2109.0 | 4.95% | motif file (matrix) |
pdf |
| 5 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-5 | -1.346e+01 | 0.0001 | 117.0 | 7.81% | 2112.6 | 4.96% | motif file (matrix) |
pdf |
| 6 |  | ETS(ETS)/Promoter/Homer | 1e-5 | -1.231e+01 | 0.0002 | 81.0 | 5.41% | 1355.7 | 3.18% | motif file (matrix) |
pdf |
| 7 |  | Fox:Ebox(Forkhead,bHLH)/Panc1-Foxa2-ChIP-Seq(GSE47459)/Homer | 1e-4 | -1.133e+01 | 0.0005 | 220.0 | 14.69% | 4727.3 | 11.09% | motif file (matrix) |
pdf |
| 8 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-4 | -1.061e+01 | 0.0008 | 170.0 | 11.35% | 3533.5 | 8.29% | motif file (matrix) |
pdf |
| 9 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-4 | -1.060e+01 | 0.0008 | 198.0 | 13.22% | 4229.2 | 9.92% | motif file (matrix) |
pdf |
| 10 |  | RXR(NR),DR1/3T3L1-RXR-ChIP-Seq(GSE13511)/Homer | 1e-4 | -9.625e+00 | 0.0017 | 193.0 | 12.88% | 4177.4 | 9.80% | motif file (matrix) |
pdf |
| 11 | 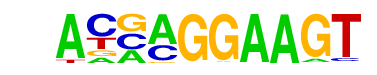 | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-4 | -9.572e+00 | 0.0017 | 144.0 | 9.61% | 2964.4 | 6.95% | motif file (matrix) |
pdf |
| 12 | 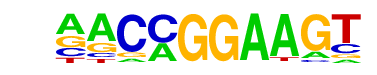 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-4 | -9.300e+00 | 0.0020 | 244.0 | 16.29% | 5504.2 | 12.91% | motif file (matrix) |
pdf |
| 13 |  | YY1(Zf)/Promoter/Homer | 1e-3 | -9.073e+00 | 0.0023 | 21.0 | 1.40% | 232.5 | 0.55% | motif file (matrix) |
pdf |
| 14 |  | AP-2alpha(AP2)/Hela-AP2alpha-ChIP-Seq(GSE31477)/Homer | 1e-3 | -8.810e+00 | 0.0028 | 163.0 | 10.88% | 3487.3 | 8.18% | motif file (matrix) |
pdf |
| 15 |  | ZFX(Zf)/mES-Zfx-ChIP-Seq(GSE11431)/Homer | 1e-3 | -8.262e+00 | 0.0045 | 229.0 | 15.29% | 5212.1 | 12.23% | motif file (matrix) |
pdf |
| 16 |  | Foxh1(Forkhead)/hESC-FOXH1-ChIP-Seq(GSE29422)/Homer | 1e-3 | -8.076e+00 | 0.0051 | 145.0 | 9.68% | 3093.7 | 7.26% | motif file (matrix) |
pdf |
| 17 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-3 | -7.874e+00 | 0.0059 | 270.0 | 18.02% | 6319.4 | 14.82% | motif file (matrix) |
pdf |
| 18 |  | AMYB(HTH)/Testes-AMYB-ChIP-Seq(GSE44588)/Homer | 1e-3 | -7.575e+00 | 0.0075 | 250.0 | 16.69% | 5827.0 | 13.67% | motif file (matrix) |
pdf |
| 19 |  | SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer | 1e-3 | -7.528e+00 | 0.0075 | 176.0 | 11.75% | 3918.8 | 9.19% | motif file (matrix) |
pdf |
| 20 |  | MYB(HTH)/ERMYB-Myb-ChIPSeq(GSE22095)/Homer | 1e-3 | -7.337e+00 | 0.0086 | 301.0 | 20.09% | 7197.7 | 16.88% | motif file (matrix) |
pdf |
| 21 |  | BMYB(HTH)/Hela-BMYB-ChIP-Seq(GSE27030)/Homer | 1e-3 | -7.326e+00 | 0.0086 | 254.0 | 16.96% | 5957.9 | 13.98% | motif file (matrix) |
pdf |
| 22 |  | E2F6(E2F)/Hela-E2F6-ChIP-Seq(GSE31477)/Homer | 1e-3 | -7.151e+00 | 0.0094 | 108.0 | 7.21% | 2245.2 | 5.27% | motif file (matrix) |
pdf |
| 23 |  | ZNF711(Zf)/SHSY5Y-ZNF711-ChIP-Seq(GSE20673)/Homer | 1e-2 | -6.830e+00 | 0.0124 | 332.0 | 22.16% | 8086.4 | 18.97% | motif file (matrix) |
pdf |
| 24 |  | Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-2 | -6.721e+00 | 0.0133 | 59.0 | 3.94% | 1100.3 | 2.58% | motif file (matrix) |
pdf |
| 25 |  | AP-2gamma(AP2)/MCF7-TFAP2C-ChIP-Seq(GSE21234)/Homer | 1e-2 | -6.677e+00 | 0.0133 | 195.0 | 13.02% | 4485.7 | 10.52% | motif file (matrix) |
pdf |
| 26 |  | PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-2 | -6.281e+00 | 0.0190 | 91.0 | 6.07% | 1888.1 | 4.43% | motif file (matrix) |
pdf |
| 27 |  | HNF4a(NR),DR1/HepG2-HNF4a-ChIP-Seq(GSE25021)/Homer | 1e-2 | -5.780e+00 | 0.0302 | 71.0 | 4.74% | 1435.8 | 3.37% | motif file (matrix) |
pdf |
| 28 |  | Smad2(MAD)/ES-SMAD2-ChIP-Seq(GSE29422)/Homer | 1e-2 | -5.763e+00 | 0.0302 | 264.0 | 17.62% | 6403.7 | 15.02% | motif file (matrix) |
pdf |
| 29 |  | AR-halfsite(NR)/LNCaP-AR-ChIP-Seq(GSE27824)/Homer | 1e-2 | -5.744e+00 | 0.0302 | 632.0 | 42.19% | 16503.2 | 38.71% | motif file (matrix) |
pdf |
| 30 |  | Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan et al.)/Homer | 1e-2 | -5.564e+00 | 0.0337 | 339.0 | 22.63% | 8446.4 | 19.81% | motif file (matrix) |
pdf |
| 31 |  | Smad4(MAD)/ESC-SMAD4-ChIP-Seq(GSE29422)/Homer | 1e-2 | -5.559e+00 | 0.0337 | 276.0 | 18.42% | 6750.6 | 15.83% | motif file (matrix) |
pdf |
| 32 |  | E2F4(E2F)/K562-E2F4-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.278e+00 | 0.0421 | 90.0 | 6.01% | 1935.3 | 4.54% | motif file (matrix) |
pdf |
| 33 |  | BMAL1(bHLH)/Liver-Bmal1-ChIP-Seq(GSE39860)/Homer | 1e-2 | -5.237e+00 | 0.0425 | 349.0 | 23.30% | 8766.4 | 20.56% | motif file (matrix) |
pdf |
| 34 |  | Foxa2(Forkhead)/Liver-Foxa2-ChIP-Seq(GSE25694)/Homer | 1e-2 | -4.902e+00 | 0.0577 | 173.0 | 11.55% | 4100.1 | 9.62% | motif file (matrix) |
pdf |
| 35 |  | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-2 | -4.773e+00 | 0.0638 | 216.0 | 14.42% | 5252.2 | 12.32% | motif file (matrix) |
pdf |
| 36 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-2 | -4.670e+00 | 0.0688 | 97.0 | 6.48% | 2160.8 | 5.07% | motif file (matrix) |
pdf |
| 37 |  | GFY-Staf(?,Zf)/Promoter/Homer | 1e-2 | -4.654e+00 | 0.0688 | 15.0 | 1.00% | 212.3 | 0.50% | motif file (matrix) |
pdf |









































































