| p-value: | 1e-4 |
| log p-value: | -1.144e+01 |
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 1.0 |
| Percentage of Target Sequences with motif | 100.00% |
| Number of Background Sequences with motif | 1.9 |
| Percentage of Background Sequences with motif | 0.00% |
| Average Position of motif in Targets | 420.0 +/- 0.0bp |
| Average Position of motif in Background | 157.0 +/- 0.0bp |
| Strand Bias (log2 ratio + to - strand density) | 10.0 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
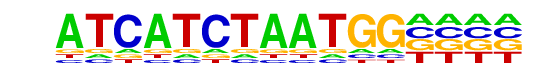
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
Isl1(Homeobox)/Neuron-Isl1-ChIP-Seq(GSE31456)/Homer
| Match Rank: | 1 |
| Score: | 0.67
| | Offset: | 5
| | Orientation: | forward strand |
| Alignment: | ATCATCTAATGG-
-----CTAATKGV |
|


|
|
MA0132.1_Pdx1/Jaspar
| Match Rank: | 2 |
| Score: | 0.62
| | Offset: | 5
| | Orientation: | forward strand |
| Alignment: | ATCATCTAATGG
-----CTAATT- |
|


|
|
Nkx6.1(Homeobox)/Islet-Nkx6.1-ChIP-Seq(GSE40975)/Homer
| Match Rank: | 3 |
| Score: | 0.59
| | Offset: | 4
| | Orientation: | forward strand |
| Alignment: | ATCATCTAATGG
----GKTAATGR |
|


|
|
Nanog(Homeobox)/mES-Nanog-ChIP-Seq(GSE11724)/Homer
| Match Rank: | 4 |
| Score: | 0.58
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | ATCATCTAATGG--
----GTTAATGGCC |
|


|
|
PH0056.1_Hoxa9/Jaspar
| Match Rank: | 5 |
| Score: | 0.56
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -ATCATCTAATGG----
ANTAATTTTATGGCCGN |
|


|
|
MA0488.1_JUN/Jaspar
| Match Rank: | 6 |
| Score: | 0.54
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---ATCATCTAATGG
AAGATGATGTCAT-- |
|


|
|
PH0037.1_Hdx/Jaspar
| Match Rank: | 7 |
| Score: | 0.54
| | Offset: | -8
| | Orientation: | forward strand |
| Alignment: | --------ATCATCTAATGG
AAGGCGAAATCATCGCA--- |
|


|
|
MA0091.1_TAL1::TCF3/Jaspar
| Match Rank: | 8 |
| Score: | 0.53
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --ATCATCTAATGG
CGACCATCTGTT-- |
|


|
|
PH0009.1_Bsx/Jaspar
| Match Rank: | 9 |
| Score: | 0.53
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | ATCATCTAATGG----
NTNAGNTAATTACCTN |
|

|
|
MA0089.1_NFE2L1::MafG/Jaspar
| Match Rank: | 10 |
| Score: | 0.53
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | ATCATCTAATGG
GTCATN------ |
|


|
|





















