| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-16 | -3.872e+01 | 0.0000 | 281.0 | 11.99% | 3163.6 | 7.09% | motif file (matrix) |
pdf |
| 2 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-16 | -3.823e+01 | 0.0000 | 285.0 | 12.16% | 3235.5 | 7.26% | motif file (matrix) |
pdf |
| 3 | 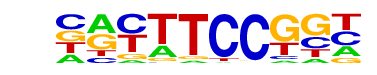 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-15 | -3.679e+01 | 0.0000 | 439.0 | 18.74% | 5676.4 | 12.73% | motif file (matrix) |
pdf |
| 4 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-12 | -2.907e+01 | 0.0000 | 245.0 | 10.46% | 2882.5 | 6.46% | motif file (matrix) |
pdf |
| 5 |  | ETS(ETS)/Promoter/Homer | 1e-12 | -2.808e+01 | 0.0000 | 168.0 | 7.17% | 1775.6 | 3.98% | motif file (matrix) |
pdf |
| 6 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-11 | -2.762e+01 | 0.0000 | 342.0 | 14.60% | 4443.8 | 9.97% | motif file (matrix) |
pdf |
| 7 | 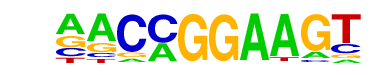 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-11 | -2.624e+01 | 0.0000 | 475.0 | 20.27% | 6686.7 | 14.99% | motif file (matrix) |
pdf |
| 8 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-10 | -2.499e+01 | 0.0000 | 220.0 | 9.39% | 2621.0 | 5.88% | motif file (matrix) |
pdf |
| 9 | 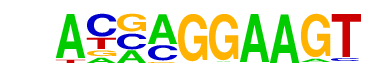 | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-8 | -2.019e+01 | 0.0000 | 245.0 | 10.46% | 3167.6 | 7.10% | motif file (matrix) |
pdf |
| 10 |  | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-8 | -1.960e+01 | 0.0000 | 410.0 | 17.50% | 5906.4 | 13.24% | motif file (matrix) |
pdf |
| 11 |  | YY1(Zf)/Promoter/Homer | 1e-8 | -1.889e+01 | 0.0000 | 52.0 | 2.22% | 399.0 | 0.89% | motif file (matrix) |
pdf |
| 12 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-7 | -1.650e+01 | 0.0000 | 494.0 | 21.08% | 7521.9 | 16.87% | motif file (matrix) |
pdf |
| 13 |  | Sox4(HMG)/proB-Sox4-ChIP-Seq(GSE50066)/Homer | 1e-7 | -1.624e+01 | 0.0000 | 239.0 | 10.20% | 3225.9 | 7.23% | motif file (matrix) |
pdf |
| 14 |  | Sox10(HMG)/SciaticNerve-Sox3-ChIP-Seq(GSE35132)/Homer | 1e-6 | -1.492e+01 | 0.0000 | 441.0 | 18.82% | 6697.3 | 15.02% | motif file (matrix) |
pdf |
| 15 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-6 | -1.444e+01 | 0.0000 | 347.0 | 14.81% | 5110.6 | 11.46% | motif file (matrix) |
pdf |
| 16 |  | EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer | 1e-6 | -1.398e+01 | 0.0000 | 216.0 | 9.22% | 2948.7 | 6.61% | motif file (matrix) |
pdf |
| 17 |  | Sox2(HMG)/mES-Sox2-ChIP-Seq(GSE11431)/Homer | 1e-5 | -1.306e+01 | 0.0000 | 239.0 | 10.20% | 3368.8 | 7.55% | motif file (matrix) |
pdf |
| 18 |  | Sox3(HMG)/NPC-Sox3-ChIP-Seq(GSE33059)/Homer | 1e-5 | -1.225e+01 | 0.0001 | 462.0 | 19.72% | 7241.9 | 16.24% | motif file (matrix) |
pdf |
| 19 |  | ZFX(Zf)/mES-Zfx-ChIP-Seq(GSE11431)/Homer | 1e-4 | -1.125e+01 | 0.0002 | 469.0 | 20.02% | 7438.8 | 16.68% | motif file (matrix) |
pdf |
| 20 |  | IRF2(IRF)/Erythroblas-IRF2-ChIP-Seq(GSE36985)/Homer | 1e-4 | -1.107e+01 | 0.0002 | 42.0 | 1.79% | 387.6 | 0.87% | motif file (matrix) |
pdf |
| 21 |  | E2F4(E2F)/K562-E2F4-ChIP-Seq(GSE31477)/Homer | 1e-4 | -1.095e+01 | 0.0002 | 211.0 | 9.01% | 3008.3 | 6.75% | motif file (matrix) |
pdf |
| 22 |  | E2F6(E2F)/Hela-E2F6-ChIP-Seq(GSE31477)/Homer | 1e-4 | -9.930e+00 | 0.0006 | 240.0 | 10.24% | 3550.5 | 7.96% | motif file (matrix) |
pdf |
| 23 |  | Sox6(HMG)/Myotubes-Sox6-ChIP-Seq(GSE32627)/Homer | 1e-4 | -9.690e+00 | 0.0007 | 410.0 | 17.50% | 6515.9 | 14.61% | motif file (matrix) |
pdf |
| 24 |  | EBF1(EBF)/Near-E2A-ChIP-Seq(GSE21512)/Homer | 1e-4 | -9.385e+00 | 0.0009 | 384.0 | 16.39% | 6080.8 | 13.64% | motif file (matrix) |
pdf |
| 25 |  | p53(p53)/Saos-p53-ChIP-Seq(GSE15780)/Homer | 1e-3 | -8.817e+00 | 0.0015 | 37.0 | 1.58% | 361.7 | 0.81% | motif file (matrix) |
pdf |
| 26 |  | p53(p53)/Saos-p53-ChIP-Seq/Homer | 1e-3 | -8.817e+00 | 0.0015 | 37.0 | 1.58% | 361.7 | 0.81% | motif file (matrix) |
pdf |
| 27 |  | E2F(E2F)/Hela-CellCycle-Expression/Homer | 1e-3 | -8.312e+00 | 0.0024 | 29.0 | 1.24% | 264.4 | 0.59% | motif file (matrix) |
pdf |
| 28 |  | PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-3 | -8.259e+00 | 0.0024 | 151.0 | 6.44% | 2149.4 | 4.82% | motif file (matrix) |
pdf |
| 29 |  | ISRE(IRF)/ThioMac-LPS-Expression(GSE23622)/Homer | 1e-3 | -7.951e+00 | 0.0032 | 28.0 | 1.20% | 257.1 | 0.58% | motif file (matrix) |
pdf |
| 30 |  | PPARE(NR),DR1/3T3L1-Pparg-ChIP-Seq(GSE13511)/Homer | 1e-3 | -7.926e+00 | 0.0032 | 280.0 | 11.95% | 4368.1 | 9.80% | motif file (matrix) |
pdf |
| 31 |  | BMYB(HTH)/Hela-BMYB-ChIP-Seq(GSE27030)/Homer | 1e-3 | -7.390e+00 | 0.0053 | 385.0 | 16.43% | 6263.5 | 14.05% | motif file (matrix) |
pdf |
| 32 |  | Foxh1(Forkhead)/hESC-FOXH1-ChIP-Seq(GSE29422)/Homer | 1e-3 | -7.343e+00 | 0.0053 | 178.0 | 7.60% | 2652.6 | 5.95% | motif file (matrix) |
pdf |
| 33 |  | E2F7(E2F)/Hela-E2F7-ChIP-Seq(GSE32673)/Homer | 1e-3 | -7.128e+00 | 0.0064 | 63.0 | 2.69% | 782.9 | 1.76% | motif file (matrix) |
pdf |
| 34 |  | RXR(NR),DR1/3T3L1-RXR-ChIP-Seq(GSE13511)/Homer | 1e-3 | -7.056e+00 | 0.0067 | 330.0 | 14.08% | 5315.9 | 11.92% | motif file (matrix) |
pdf |
| 35 |  | AMYB(HTH)/Testes-AMYB-ChIP-Seq(GSE44588)/Homer | 1e-3 | -7.047e+00 | 0.0067 | 394.0 | 16.82% | 6455.2 | 14.48% | motif file (matrix) |
pdf |
| 36 |  | HNF4a(NR),DR1/HepG2-HNF4a-ChIP-Seq(GSE25021)/Homer | 1e-2 | -6.740e+00 | 0.0087 | 122.0 | 5.21% | 1746.1 | 3.92% | motif file (matrix) |
pdf |
| 37 |  | Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-2 | -6.569e+00 | 0.0100 | 91.0 | 3.88% | 1246.0 | 2.79% | motif file (matrix) |
pdf |
| 38 |  | PRDM1(Zf)/Hela-PRDM1-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.491e+00 | 0.0105 | 148.0 | 6.32% | 2195.8 | 4.92% | motif file (matrix) |
pdf |
| 39 |  | ZNF711(Zf)/SHSY5Y-ZNF711-ChIP-Seq(GSE20673)/Homer | 1e-2 | -6.439e+00 | 0.0108 | 674.0 | 28.77% | 11615.6 | 26.05% | motif file (matrix) |
pdf |
| 40 |  | MYB(HTH)/ERMYB-Myb-ChIPSeq(GSE22095)/Homer | 1e-2 | -6.093e+00 | 0.0149 | 457.0 | 19.50% | 7686.4 | 17.24% | motif file (matrix) |
pdf |
| 41 |  | Tcfcp2l1(CP2)/mES-Tcfcp2l1-ChIP-Seq(GSE11431)/Homer | 1e-2 | -5.894e+00 | 0.0177 | 44.0 | 1.88% | 531.4 | 1.19% | motif file (matrix) |
pdf |
| 42 |  | SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer | 1e-2 | -5.777e+00 | 0.0195 | 280.0 | 11.95% | 4541.4 | 10.18% | motif file (matrix) |
pdf |
| 43 |  | PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-2 | -5.580e+00 | 0.0232 | 367.0 | 15.66% | 6115.9 | 13.71% | motif file (matrix) |
pdf |
| 44 |  | Fox:Ebox(Forkhead,bHLH)/Panc1-Foxa2-ChIP-Seq(GSE47459)/Homer | 1e-2 | -5.441e+00 | 0.0260 | 266.0 | 11.35% | 4323.1 | 9.69% | motif file (matrix) |
pdf |
| 45 |  | TR4(NR),DR1/Hela-TR4-ChIP-Seq(GSE24685)/Homer | 1e-2 | -5.331e+00 | 0.0284 | 35.0 | 1.49% | 412.5 | 0.93% | motif file (matrix) |
pdf |
| 46 |  | HIF-1b(HLH)/T47D-HIF1b-ChIP-Seq(GSE59937)/Homer | 1e-2 | -5.318e+00 | 0.0284 | 374.0 | 15.96% | 6269.8 | 14.06% | motif file (matrix) |
pdf |
| 47 | 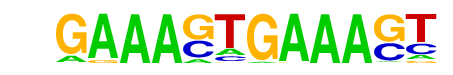 | IRF1(IRF)/PBMC-IRF1-ChIP-Seq(GSE43036)/Homer | 1e-2 | -5.314e+00 | 0.0284 | 46.0 | 1.96% | 580.6 | 1.30% | motif file (matrix) |
pdf |
| 48 |  | AR-halfsite(NR)/LNCaP-AR-ChIP-Seq(GSE27824)/Homer | 1e-2 | -5.256e+00 | 0.0287 | 1040.0 | 44.39% | 18618.4 | 41.75% | motif file (matrix) |
pdf |
| 49 |  | E2F1(E2F)/Hela-E2F1-ChIP-Seq(GSE22478)/Homer | 1e-2 | -5.176e+00 | 0.0305 | 117.0 | 4.99% | 1749.9 | 3.92% | motif file (matrix) |
pdf |
| 50 |  | TATA-Box(TBP)/Promoter/Homer | 1e-2 | -5.140e+00 | 0.0309 | 317.0 | 13.53% | 5262.7 | 11.80% | motif file (matrix) |
pdf |
| 51 |  | IRF4(IRF)/GM12878-IRF4-ChIP-Seq(GSE32465)/Homer | 1e-2 | -4.852e+00 | 0.0404 | 108.0 | 4.61% | 1616.7 | 3.63% | motif file (matrix) |
pdf |
| 52 |  | ETS:RUNX(ETS,Runt)/Jurkat-RUNX1-ChIP-Seq(GSE17954)/Homer | 1e-2 | -4.825e+00 | 0.0408 | 39.0 | 1.66% | 488.3 | 1.10% | motif file (matrix) |
pdf |
| 53 |  | AP-2alpha(AP2)/Hela-AP2alpha-ChIP-Seq(GSE31477)/Homer | 1e-2 | -4.704e+00 | 0.0451 | 316.0 | 13.49% | 5290.9 | 11.86% | motif file (matrix) |
pdf |









































































































