| p-value: | 1e-18 |
| log p-value: | -4.341e+01 |
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 10.0 |
| Percentage of Target Sequences with motif | 3.60% |
| Number of Background Sequences with motif | 11.0 |
| Percentage of Background Sequences with motif | 0.02% |
| Average Position of motif in Targets | 199.8 +/- 99.3bp |
| Average Position of motif in Background | 147.6 +/- 79.7bp |
| Strand Bias (log2 ratio + to - strand density) | 3.5 |
| Multiplicity (# of sites on avg that occur together) | 1.20 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PB0146.1_Mafk_2/Jaspar
| Match Rank: | 1 |
| Score: | 0.62
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --AGAATCTGCAAG-
GAAAAAATTGCAAGG |
|


|
|
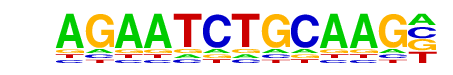
IRF1(IRF)/PBMC-IRF1-ChIP-Seq(GSE43036)/Homer
| Match Rank: | 2 |
| Score: | 0.61
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | AGAATCTGCAAG-
-GAAAGTGAAAGT |
|

|
|
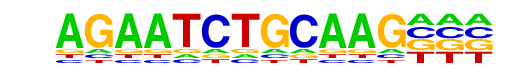
MA0486.1_HSF1/Jaspar
| Match Rank: | 3 |
| Score: | 0.59
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | AGAATCTGCAAG---
AGAANNTTCTAGAAN |
|

|
|
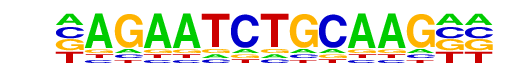
HRE(HSF)/Striatum-HSF1-ChIP-Seq(GSE38000)/Homer
| Match Rank: | 4 |
| Score: | 0.59
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -AGAATCTGCAAG--
TAGAANVTTCTAGAA |
|

|
|
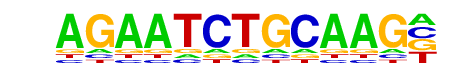
ISRE(IRF)/ThioMac-LPS-Expression(GSE23622)/Homer
| Match Rank: | 5 |
| Score: | 0.58
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | AGAATCTGCAAG-
-GAAACTGAAACT |
|

|
|
HRE(HSF)/HepG2-HSF1-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 6 |
| Score: | 0.58
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----AGAATCTGCAAG----
TTCTAGAANNTTCCAGAANN |
|


|
|
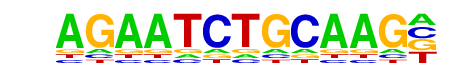
IRF2(IRF)/Erythroblas-IRF2-ChIP-Seq(GSE36985)/Homer
| Match Rank: | 7 |
| Score: | 0.57
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | AGAATCTGCAAG-
-GAAASYGAAASY |
|

|
|
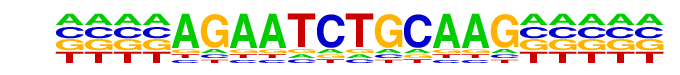
MA0050.2_IRF1/Jaspar
| Match Rank: | 8 |
| Score: | 0.56
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----AGAATCTGCAAG-----
AAANNGAAAGTGAAAGTAAAN |
|

|
|
Pit1(Homeobox)/GCrat-Pit1-ChIP-Seq(GSE58009)/Homer
| Match Rank: | 9 |
| Score: | 0.55
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | AGAATCTGCAAG
-GHATATKCAT- |
|


|
|
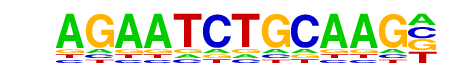
Oct2(POU,Homeobox)/Bcell-Oct2-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 10 |
| Score: | 0.55
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | AGAATCTGCAAG-
---ATATGCAAAT |
|

|
|





















