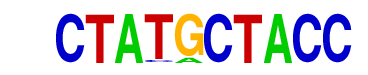
| p-value: | 1e-7 |
| log p-value: | -1.692e+01 |
| Information Content per bp: | 1.957 |
| Number of Target Sequences with motif | 7.0 |
| Percentage of Target Sequences with motif | 1.23% |
| Number of Background Sequences with motif | 27.7 |
| Percentage of Background Sequences with motif | 0.06% |
| Average Position of motif in Targets | 218.3 +/- 138.1bp |
| Average Position of motif in Background | 230.5 +/- 114.0bp |
| Strand Bias (log2 ratio + to - strand density) | 2.5 |
| Multiplicity (# of sites on avg that occur together) | 1.86 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PB0154.1_Osr1_2/Jaspar
| Match Rank: | 1 |
| Score: | 0.70
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CTATGCTACC------
ACATGCTACCTAATAC |
|


|
|
PB0155.1_Osr2_2/Jaspar
| Match Rank: | 2 |
| Score: | 0.64
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CTATGCTACC------
ACTTGCTACCTACACC |
|


|
|
PB0051.1_Osr2_1/Jaspar
| Match Rank: | 3 |
| Score: | 0.64
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CTATGCTACC------
CNNNGCTACTGTANNN |
|


|
|
PB0050.1_Osr1_1/Jaspar
| Match Rank: | 4 |
| Score: | 0.62
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CTATGCTACC------
TNNTGCTACTGTNNNN |
|


|
|
PB0059.1_Six6_1/Jaspar
| Match Rank: | 5 |
| Score: | 0.61
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CTATGCTACC-----
ANANNTGATACCCNATN |
|


|
|
PB0029.1_Hic1_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.60
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CTATGCTACC-----
ACTATGCCAACCTACC |
|


|
|
PH0161.1_Six1/Jaspar
| Match Rank: | 7 |
| Score: | 0.59
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CTATGCTACC-----
ANNNATGATACCCCATC |
|


|
|
PH0166.1_Six6_2/Jaspar
| Match Rank: | 8 |
| Score: | 0.57
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CTATGCTACC-----
AATNTTGATACCCTATN |
|


|
|
EBNA1(EBV virus)/Raji-EBNA1-ChIP-Seq(GSE30709)/Homer
| Match Rank: | 9 |
| Score: | 0.55
| | Offset: | -10
| | Orientation: | reverse strand |
| Alignment: | ----------CTATGCTACC
NNNGGGHAGCAHRTGCTRCC |
|


|
|
PH0165.1_Six6_1/Jaspar
| Match Rank: | 10 |
| Score: | 0.55
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CTATGCTACC-----
ANANNTGATACCCTATN |
|


|
|