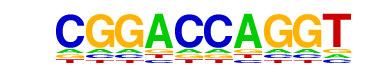
| p-value: | 1e-2 |
| log p-value: | -6.764e+00 |
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 1.0 |
| Percentage of Target Sequences with motif | 4.76% |
| Number of Background Sequences with motif | 2.1 |
| Percentage of Background Sequences with motif | 0.01% |
| Average Position of motif in Targets | 263.0 +/- 0.0bp |
| Average Position of motif in Background | 308.2 +/- 110.3bp |
| Strand Bias (log2 ratio + to - strand density) | 10.0 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PB0200.1_Zfp187_2/Jaspar
| Match Rank: | 1 |
| Score: | 0.72
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---CGGACCAGGT---
NNAGGGACAAGGGCNC |
|
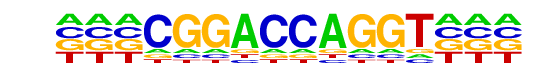

|
|
E2A(bHLH),near_PU.1/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 2 |
| Score: | 0.64
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | CGGACCAGGT---
---NNCAGGTGNN |
|


|
|
MA0103.2_ZEB1/Jaspar
| Match Rank: | 3 |
| Score: | 0.62
| | Offset: | 5
| | Orientation: | reverse strand |
| Alignment: | CGGACCAGGT----
-----CAGGTGAGG |
|


|
|
MF0007.1_bHLH(zip)_class/Jaspar
| Match Rank: | 4 |
| Score: | 0.58
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | CGGACCAGGT-
---ACCACGTG |
|


|
|
MA0059.1_MYC::MAX/Jaspar
| Match Rank: | 5 |
| Score: | 0.57
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | CGGACCAGGT----
---ACCACGTGCTC |
|


|
|
PB0043.1_Max_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.57
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CGGACCAGGT-------
-TGACCACGTGGTCGGG |
|


|
|
PB0089.1_Tcfe2a_1/Jaspar
| Match Rank: | 7 |
| Score: | 0.57
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CGGACCAGGT-------
ATCCACAGGTGCGAAAA |
|


|
|
POL013.1_MED-1/Jaspar
| Match Rank: | 8 |
| Score: | 0.57
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | CGGACCAGGT
CGGAGC---- |
|
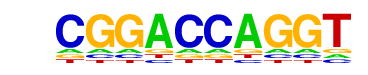

|
|
PB0047.1_Myf6_1/Jaspar
| Match Rank: | 9 |
| Score: | 0.56
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -CGGACCAGGT-----
GAAGAACAGGTGTCCG |
|


|
|
MA0104.3_Mycn/Jaspar
| Match Rank: | 10 |
| Score: | 0.55
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | CGGACCAGGT-
---GCCACGTG |
|


|
|