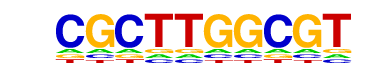
| p-value: | 1e-3 |
| log p-value: | -7.457e+00 |
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 1.0 |
| Percentage of Target Sequences with motif | 4.76% |
| Number of Background Sequences with motif | 1.6 |
| Percentage of Background Sequences with motif | 0.00% |
| Average Position of motif in Targets | 104.0 +/- 0.0bp |
| Average Position of motif in Background | 135.6 +/- 87.4bp |
| Strand Bias (log2 ratio + to - strand density) | 10.0 |
| Multiplicity (# of sites on avg that occur together) | 1.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
MA0161.1_NFIC/Jaspar
| Match Rank: | 1 |
| Score: | 0.66
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | CGCTTGGCGT
---TTGGCA- |
|

|
|
PB0113.1_E2F3_2/Jaspar
| Match Rank: | 2 |
| Score: | 0.61
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CGCTTGGCGT------
NNNNTTGGCGCCGANNN |
|


|
|
PB0112.1_E2F2_2/Jaspar
| Match Rank: | 3 |
| Score: | 0.60
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CGCTTGGCGT------
NNNNTTGGCGCCGANNN |
|


|
|
PB0147.1_Max_2/Jaspar
| Match Rank: | 4 |
| Score: | 0.60
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----CGCTTGGCGT
NNGTCGCGTGNCAC |
|


|
|
MF0002.1_bZIP_CREB/G-box-like_subclass/Jaspar
| Match Rank: | 5 |
| Score: | 0.59
| | Offset: | 4
| | Orientation: | forward strand |
| Alignment: | CGCTTGGCGT
----TGACGT |
|

|
|
PB0029.1_Hic1_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.58
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---CGCTTGGCGT---
NGTAGGTTGGCATNNN |
|

|
|
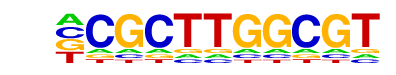
NFY(CCAAT)/Promoter/Homer
| Match Rank: | 7 |
| Score: | 0.57
| | Offset: | -1
| | Orientation: | reverse strand |
| Alignment: | -CGCTTGGCGT
CCGATTGGCT- |
|

|
|
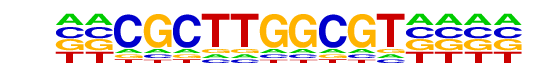
PB0032.1_IRC900814_1/Jaspar
| Match Rank: | 8 |
| Score: | 0.55
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --CGCTTGGCGT----
GNNATTTGTCGTAANN |
|

|
|
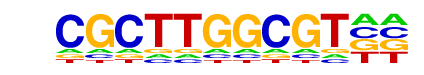
MA0018.2_CREB1/Jaspar
| Match Rank: | 9 |
| Score: | 0.54
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | CGCTTGGCGT--
----TGACGTCA |
|

|
|
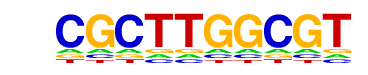
MA0006.1_Arnt::Ahr/Jaspar
| Match Rank: | 10 |
| Score: | 0.54
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CGCTTGGCGT
TGCGTG---- |
|

|
|