| p-value: | 1e-3 |
| log p-value: | -8.206e+00 |
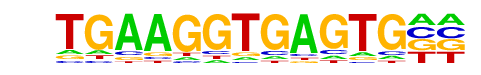
| Information Content per bp: | 1.584 |
| Number of Target Sequences with motif | 4.0 |
| Percentage of Target Sequences with motif | 0.09% |
| Number of Background Sequences with motif | 3.6 |
| Percentage of Background Sequences with motif | 0.01% |
| Average Position of motif in Targets | 189.0 +/- 101.4bp |
| Average Position of motif in Background | 245.1 +/- 34.9bp |
| Strand Bias (log2 ratio + to - strand density) | 1.8 |
| Multiplicity (# of sites on avg that occur together) | 2.25 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
SD0001.1_at_AC_acceptor/Jaspar
| Match Rank: | 1 |
| Score: | 0.74
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | TGAAGGTGAGTG-
--CAGGTAAGTAT |
|


|
|
Esrrb(NR)/mES-Esrrb-ChIP-Seq(GSE11431)/Homer
| Match Rank: | 2 |
| Score: | 0.64
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TGAAGGTGAGTG
TCAAGGTCAN-- |
|


|
|
MA0103.2_ZEB1/Jaspar
| Match Rank: | 3 |
| Score: | 0.63
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | TGAAGGTGAGTG
--CAGGTGAGG- |
|


|
|
MA0141.2_Esrrb/Jaspar
| Match Rank: | 4 |
| Score: | 0.63
| | Offset: | -3
| | Orientation: | forward strand |
| Alignment: | ---TGAAGGTGAGTG
AGCTCAAGGTCA--- |
|


|
|
MA0503.1_Nkx2-5_(var.2)/Jaspar
| Match Rank: | 5 |
| Score: | 0.60
| | Offset: | 4
| | Orientation: | reverse strand |
| Alignment: | TGAAGGTGAGTG---
----CTTGAGTGGCT |
|


|
|
MA0071.1_RORA_1/Jaspar
| Match Rank: | 6 |
| Score: | 0.60
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -TGAAGGTGAGTG
ATCAAGGTCA--- |
|


|
|
Nr5a2(NR)/Pancreas-LRH1-ChIP-Seq(GSE34295)/Homer
| Match Rank: | 7 |
| Score: | 0.58
| | Offset: | -1
| | Orientation: | forward strand |
| Alignment: | -TGAAGGTGAGTG
BTCAAGGTCA--- |
|


|
|
MA0122.1_Nkx3-2/Jaspar
| Match Rank: | 8 |
| Score: | 0.58
| | Offset: | 5
| | Orientation: | forward strand |
| Alignment: | TGAAGGTGAGTG--
-----TTAAGTGGA |
|

|
|
Nur77(NR)/K562-NR4A1-ChIP-Seq(GSE31363)/Homer
| Match Rank: | 9 |
| Score: | 0.58
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---TGAAGGTGAGTG
ANGNAAAGGTCA--- |
|


|
|
E2A(bHLH),near_PU.1/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 10 |
| Score: | 0.58
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | TGAAGGTGAGTG
NNCAGGTGNN-- |
|


|
|





















