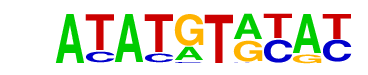
| p-value: | 1e-4 |
| log p-value: | -9.448e+00 |
| Information Content per bp: | 1.755 |
| Number of Target Sequences with motif | 78.0 |
| Percentage of Target Sequences with motif | 6.79% |
| Number of Background Sequences with motif | 2090.6 |
| Percentage of Background Sequences with motif | 4.32% |
| Average Position of motif in Targets | 257.5 +/- 139.6bp |
| Average Position of motif in Background | 244.5 +/- 138.0bp |
| Strand Bias (log2 ratio + to - strand density) | 0.2 |
| Multiplicity (# of sites on avg that occur together) | 1.95 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
PB0198.1_Zfp128_2/Jaspar
| Match Rank: | 1 |
| Score: | 0.75
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---ATATGTRYAY-
NNTATANATATACN |
|


|
|
PB0163.1_Six6_2/Jaspar
| Match Rank: | 2 |
| Score: | 0.71
| | Offset: | -5
| | Orientation: | forward strand |
| Alignment: | -----ATATGTRYAY--
ATGGGATATATCCGCCT |
|


|
|
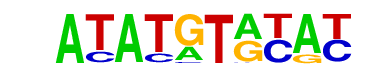
MA0033.1_FOXL1/Jaspar
| Match Rank: | 3 |
| Score: | 0.70
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | ATATGTRYAY
-TATGTNTA- |
|

|
|
PH0086.1_Irx5/Jaspar
| Match Rank: | 4 |
| Score: | 0.69
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------ATATGTRYAY-
ANTNNTACATGTANNTN |
|


|
|
PH0082.1_Irx2/Jaspar
| Match Rank: | 5 |
| Score: | 0.69
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------ATATGTRYAY-
ANTNTTACATGTATNTA |
|


|
|
PH0084.1_Irx3_2/Jaspar
| Match Rank: | 6 |
| Score: | 0.69
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------ATATGTRYAY-
NNTATTACATGTANNNT |
|


|
|
PH0087.1_Irx6/Jaspar
| Match Rank: | 7 |
| Score: | 0.68
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------ATATGTRYAY-
ANTTNTACATGTANTTN |
|


|
|
PH0083.1_Irx3_1/Jaspar
| Match Rank: | 8 |
| Score: | 0.68
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------ATATGTRYAY-
ANTATTACATGTANNNN |
|


|
|
PH0085.1_Irx4/Jaspar
| Match Rank: | 9 |
| Score: | 0.67
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------ATATGTRYAY-
NNTTTTACATGTANNNT |
|


|
|
PB0104.1_Zscan4_1/Jaspar
| Match Rank: | 10 |
| Score: | 0.67
| | Offset: | -2
| | Orientation: | reverse strand |
| Alignment: | --ATATGTRYAY-----
NTNTATGTGCACATNNN |
|


|
|