| p-value: | 1e-4 |
| log p-value: | -9.837e+00 |
| Information Content per bp: | 1.944 |
| Number of Target Sequences with motif | 4.0 |
| Percentage of Target Sequences with motif | 22.22% |
| Number of Background Sequences with motif | 471.9 |
| Percentage of Background Sequences with motif | 1.19% |
| Average Position of motif in Targets | 373.8 +/- 178.9bp |
| Average Position of motif in Background | 275.5 +/- 290.4bp |
| Strand Bias (log2 ratio + to - strand density) | -2.3 |
| Multiplicity (# of sites on avg that occur together) | 1.25 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
MA0464.1_Bhlhe40/Jaspar
| Match Rank: | 1 |
| Score: | 0.72
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | CATGCATGTG--
-NTGCACGTGAG |
|


|
|
PB0170.1_Sox17_2/Jaspar
| Match Rank: | 2 |
| Score: | 0.70
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----CATGCATGTG---
NTTNTATGAATGTGNNC |
|


|
|
MA0058.2_MAX/Jaspar
| Match Rank: | 3 |
| Score: | 0.70
| | Offset: | 3
| | Orientation: | reverse strand |
| Alignment: | CATGCATGTG---
---CCATGTGCTT |
|


|
|
PB0178.1_Sox8_2/Jaspar
| Match Rank: | 4 |
| Score: | 0.68
| | Offset: | -5
| | Orientation: | reverse strand |
| Alignment: | -----CATGCATGTG
NNTNTCATGAATGT- |
|


|
|
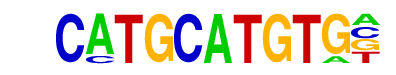
Pit1(Homeobox)/GCrat-Pit1-ChIP-Seq(GSE58009)/Homer
| Match Rank: | 5 |
| Score: | 0.67
| | Offset: | 1
| | Orientation: | forward strand |
| Alignment: | CATGCATGTG-
-ATGMATATDC |
|

|
|
MA0147.2_Myc/Jaspar
| Match Rank: | 6 |
| Score: | 0.66
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | CATGCATGTG---
---CCATGTGCTT |
|


|
|
bHLHE40(bHLH)/HepG2-BHLHE40-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 7 |
| Score: | 0.64
| | Offset: | 3
| | Orientation: | forward strand |
| Alignment: | CATGCATGTG---
---KCACGTGMCN |
|


|
|
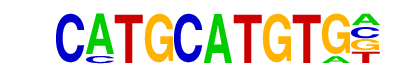
SD0002.1_at_AC_acceptor/Jaspar
| Match Rank: | 8 |
| Score: | 0.63
| | Offset: | 0
| | Orientation: | forward strand |
| Alignment: | CATGCATGTG-
AAGGCAAGTGT |
|

|
|
NPAS2(bHLH)/Liver-NPAS2-ChIP-Seq(GSE39860)/Homer
| Match Rank: | 9 |
| Score: | 0.62
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | CATGCATGTG--
--KCCACGTGAC |
|


|
|
E2A(bHLH),near_PU.1/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer
| Match Rank: | 10 |
| Score: | 0.62
| | Offset: | 2
| | Orientation: | reverse strand |
| Alignment: | CATGCATGTG--
--NNCAGGTGNN |
|


|
|





















