| p-value: | 1e-2 |
| log p-value: | -5.214e+00 |
| Information Content per bp: | 1.530 |
| Number of Target Sequences with motif | 3.0 |
| Percentage of Target Sequences with motif | 20.00% |
| Number of Background Sequences with motif | 892.7 |
| Percentage of Background Sequences with motif | 2.46% |
| Average Position of motif in Targets | 213.5 +/- 155.2bp |
| Average Position of motif in Background | 238.6 +/- 180.0bp |
| Strand Bias (log2 ratio + to - strand density) | 10.0 |
| Multiplicity (# of sites on avg that occur together) | 2.00 |
| Motif File: | file (matrix)
reverse opposite |
| PDF Format Logos: | forward logo
reverse opposite |
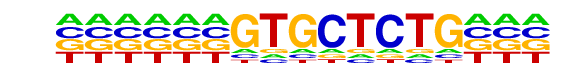
PB0099.1_Zfp691_1/Jaspar
| Match Rank: | 1 |
| Score: | 0.70
| | Offset: | -6
| | Orientation: | forward strand |
| Alignment: | ------GTGCTCTG---
CGAACAGTGCTCACTAT |
|

|
|
POL013.1_MED-1/Jaspar
| Match Rank: | 2 |
| Score: | 0.64
| | Offset: | 2
| | Orientation: | forward strand |
| Alignment: | GTGCTCTG
--GCTCCG |
|


|
|
POL010.1_DCE_S_III/Jaspar
| Match Rank: | 3 |
| Score: | 0.61
| | Offset: | 1
| | Orientation: | reverse strand |
| Alignment: | GTGCTCTG
-NGCTN-- |
|


|
|
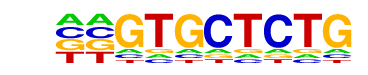
Znf263(Zf)/K562-Znf263-ChIP-Seq(GSE31477)/Homer
| Match Rank: | 4 |
| Score: | 0.60
| | Offset: | -2
| | Orientation: | forward strand |
| Alignment: | --GTGCTCTG
CNGTCCTCCC |
|

|
|
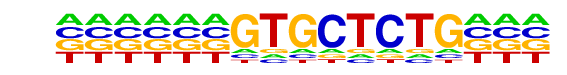
PB0196.1_Zbtb7b_2/Jaspar
| Match Rank: | 5 |
| Score: | 0.60
| | Offset: | -6
| | Orientation: | reverse strand |
| Alignment: | ------GTGCTCTG---
NNANTGGTGGTCTTNNN |
|

|
|
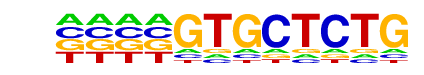
Nkx3.1(Homeobox)/LNCaP-Nkx3.1-ChIP-Seq(GSE28264)/Homer
| Match Rank: | 6 |
| Score: | 0.59
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----GTGCTCTG
TTAAGTGCTT-- |
|

|
|
MA0160.1_NR4A2/Jaspar
| Match Rank: | 7 |
| Score: | 0.57
| | Offset: | 0
| | Orientation: | reverse strand |
| Alignment: | GTGCTCTG
GTGACCTT |
|


|
|
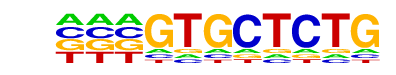
MA0512.1_Rxra/Jaspar
| Match Rank: | 8 |
| Score: | 0.57
| | Offset: | -3
| | Orientation: | reverse strand |
| Alignment: | ---GTGCTCTG
NCTGACCTTTG |
|

|
|
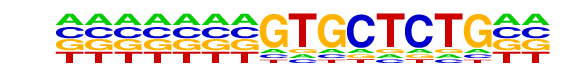
PB0152.1_Nkx3-1_2/Jaspar
| Match Rank: | 9 |
| Score: | 0.56
| | Offset: | -7
| | Orientation: | forward strand |
| Alignment: | -------GTGCTCTG--
ACTCCAAGTACTTGGAA |
|

|
|
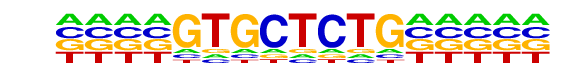
PB0091.1_Zbtb3_1/Jaspar
| Match Rank: | 10 |
| Score: | 0.56
| | Offset: | -4
| | Orientation: | reverse strand |
| Alignment: | ----GTGCTCTG-----
NNNANTGCAGTGCNNTT |
|

|
|





















