| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 |  | CTCF(Zf)/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-17 | -4.061e+01 | 0.0000 | 74.0 | 4.30% | 579.2 | 1.30% | motif file (matrix) |
pdf |
| 2 |  | BORIS(Zf)/K562-CTCFL-ChIP-Seq(GSE32465)/Homer | 1e-11 | -2.635e+01 | 0.0000 | 86.0 | 5.00% | 971.9 | 2.17% | motif file (matrix) |
pdf |
| 3 |  | GFY-Staf(?,Zf)/Promoter/Homer | 1e-4 | -9.841e+00 | 0.0047 | 25.0 | 1.45% | 263.2 | 0.59% | motif file (matrix) |
pdf |
| 4 |  | Sp1(Zf)/Promoter/Homer | 1e-4 | -9.773e+00 | 0.0047 | 93.0 | 5.40% | 1582.1 | 3.54% | motif file (matrix) |
pdf |
| 5 |  | ETS(ETS)/Promoter/Homer | 1e-4 | -9.266e+00 | 0.0050 | 98.0 | 5.69% | 1712.1 | 3.83% | motif file (matrix) |
pdf |
| 6 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-3 | -8.420e+00 | 0.0097 | 139.0 | 8.08% | 2661.7 | 5.95% | motif file (matrix) |
pdf |
| 7 |  | NRF(NRF)/Promoter/Homer | 1e-3 | -7.959e+00 | 0.0132 | 54.0 | 3.14% | 850.2 | 1.90% | motif file (matrix) |
pdf |
| 8 |  | RUNX2(Runt)/PCa-RUNX2-ChIP-Seq(GSE33889)/Homer | 1e-3 | -6.967e+00 | 0.0311 | 311.0 | 18.07% | 6837.5 | 15.29% | motif file (matrix) |
pdf |
| 9 |  | NRF1(NRF)/MCF7-NRF1-ChIP-Seq(Unpublished)/Homer | 1e-3 | -6.930e+00 | 0.0311 | 36.0 | 2.09% | 528.1 | 1.18% | motif file (matrix) |
pdf |
| 10 |  | SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer | 1e-2 | -6.714e+00 | 0.0320 | 289.0 | 16.79% | 6332.5 | 14.16% | motif file (matrix) |
pdf |
| 11 |  | CRE(bZIP)/Promoter/Homer | 1e-2 | -6.557e+00 | 0.0341 | 64.0 | 3.72% | 1117.1 | 2.50% | motif file (matrix) |
pdf |
| 12 |  | STAT5(Stat)/mCD4+-Stat5-ChIP-Seq(GSE12346)/Homer | 1e-2 | -6.542e+00 | 0.0341 | 113.0 | 6.57% | 2199.3 | 4.92% | motif file (matrix) |
pdf |
| 13 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.302e+00 | 0.0372 | 144.0 | 8.37% | 2928.0 | 6.55% | motif file (matrix) |
pdf |
| 14 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-2 | -6.004e+00 | 0.0465 | 143.0 | 8.31% | 2928.9 | 6.55% | motif file (matrix) |
pdf |
| 15 | 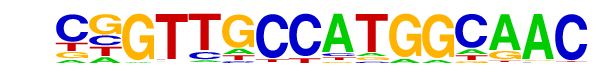 | RFX(HTH)/K562-RFX3-ChIP-Seq(SRA012198)/Homer | 1e-2 | -5.998e+00 | 0.0465 | 24.0 | 1.39% | 326.9 | 0.73% | motif file (matrix) |
pdf |
| 16 |  | CTCF-SatelliteElement(Zf?)/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-2 | -5.917e+00 | 0.0465 | 9.0 | 0.52% | 74.8 | 0.17% | motif file (matrix) |
pdf |
| 17 |  | NFY(CCAAT)/Promoter/Homer | 1e-2 | -5.486e+00 | 0.0644 | 233.0 | 13.54% | 5116.8 | 11.44% | motif file (matrix) |
pdf |
| 18 |  | E2F4(E2F)/K562-E2F4-ChIP-Seq(GSE31477)/Homer | 1e-2 | -5.244e+00 | 0.0774 | 103.0 | 5.98% | 2064.7 | 4.62% | motif file (matrix) |
pdf |
| 19 |  | E2F7(E2F)/Hela-E2F7-ChIP-Seq(GSE32673)/Homer | 1e-2 | -4.991e+00 | 0.0944 | 30.0 | 1.74% | 474.2 | 1.06% | motif file (matrix) |
pdf |
| 20 |  | STAT6(Stat)/Macrophage-Stat6-ChIP-Seq(GSE38377)/Homer | 1e-2 | -4.887e+00 | 0.0996 | 171.0 | 9.94% | 3692.2 | 8.26% | motif file (matrix) |
pdf |







































