| Rank | Motif | Name | P-value | log P-pvalue | q-value (Benjamini) | # Target Sequences with Motif | % of Targets Sequences with Motif | # Background Sequences with Motif | % of Background Sequences with Motif | Motif File |
PDF |
| 1 | 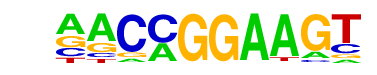 | ETV1(ETS)/GIST48-ETV1-ChIP-Seq(GSE22441)/Homer | 1e-102 | -2.353e+02 | 0.0000 | 6613.0 | 25.26% | 5117.0 | 19.80% | motif file (matrix) |
pdf |
| 2 | 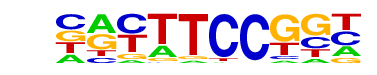 | Fli1(ETS)/CD8-FLI-ChIP-Seq(GSE20898)/Homer | 1e-96 | -2.214e+02 | 0.0000 | 5876.0 | 22.45% | 4494.1 | 17.39% | motif file (matrix) |
pdf |
| 3 |  | ERG(ETS)/VCaP-ERG-ChIP-Seq(GSE14097)/Homer | 1e-81 | -1.874e+02 | 0.0000 | 7059.0 | 26.97% | 5670.8 | 21.94% | motif file (matrix) |
pdf |
| 4 |  | GABPA(ETS)/Jurkat-GABPa-ChIP-Seq(GSE17954)/Homer | 1e-79 | -1.832e+02 | 0.0000 | 4612.0 | 17.62% | 3479.8 | 13.46% | motif file (matrix) |
pdf |
| 5 | 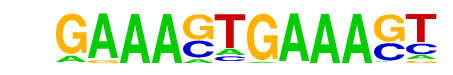 | IRF1(IRF)/PBMC-IRF1-ChIP-Seq(GSE43036)/Homer | 1e-79 | -1.820e+02 | 0.0000 | 912.0 | 3.48% | 452.4 | 1.75% | motif file (matrix) |
pdf |
| 6 |  | IRF2(IRF)/Erythroblas-IRF2-ChIP-Seq(GSE36985)/Homer | 1e-75 | -1.743e+02 | 0.0000 | 744.0 | 2.84% | 345.2 | 1.34% | motif file (matrix) |
pdf |
| 7 |  | BMYB(HTH)/Hela-BMYB-ChIP-Seq(GSE27030)/Homer | 1e-73 | -1.686e+02 | 0.0000 | 5548.0 | 21.19% | 4357.9 | 16.86% | motif file (matrix) |
pdf |
| 8 |  | ZFX(Zf)/mES-Zfx-ChIP-Seq(GSE11431)/Homer | 1e-70 | -1.622e+02 | 0.0000 | 7178.0 | 27.42% | 5868.6 | 22.71% | motif file (matrix) |
pdf |
| 9 |  | ISRE(IRF)/ThioMac-LPS-Expression(GSE23622)/Homer | 1e-64 | -1.480e+02 | 0.0000 | 471.0 | 1.80% | 190.2 | 0.74% | motif file (matrix) |
pdf |
| 10 |  | EHF(ETS)/LoVo-EHF-ChIP-Seq(GSE49402)/Homer | 1e-63 | -1.471e+02 | 0.0000 | 5602.0 | 21.40% | 4477.6 | 17.32% | motif file (matrix) |
pdf |
| 11 |  | Elk1(ETS)/Hela-Elk1-ChIP-Seq(GSE31477)/Homer | 1e-63 | -1.471e+02 | 0.0000 | 3645.0 | 13.92% | 2731.5 | 10.57% | motif file (matrix) |
pdf |
| 12 |  | ETS1(ETS)/Jurkat-ETS1-ChIP-Seq(GSE17954)/Homer | 1e-61 | -1.417e+02 | 0.0000 | 4938.0 | 18.86% | 3895.4 | 15.07% | motif file (matrix) |
pdf |
| 13 |  | Elk4(ETS)/Hela-Elk4-ChIP-Seq(GSE31477)/Homer | 1e-58 | -1.343e+02 | 0.0000 | 3625.0 | 13.85% | 2750.2 | 10.64% | motif file (matrix) |
pdf |
| 14 |  | MYB(HTH)/ERMYB-Myb-ChIPSeq(GSE22095)/Homer | 1e-53 | -1.240e+02 | 0.0000 | 6596.0 | 25.20% | 5477.5 | 21.19% | motif file (matrix) |
pdf |
| 15 |  | AMYB(HTH)/Testes-AMYB-ChIP-Seq(GSE44588)/Homer | 1e-53 | -1.233e+02 | 0.0000 | 5620.0 | 21.47% | 4580.9 | 17.72% | motif file (matrix) |
pdf |
| 16 |  | ELF1(ETS)/Jurkat-ELF1-ChIP-Seq(SRA014231)/Homer | 1e-52 | -1.216e+02 | 0.0000 | 3317.0 | 12.67% | 2517.1 | 9.74% | motif file (matrix) |
pdf |
| 17 |  | ELF5(ETS)/T47D-ELF5-ChIP-Seq(GSE30407)/Homer | 1e-50 | -1.170e+02 | 0.0000 | 3329.0 | 12.72% | 2541.2 | 9.83% | motif file (matrix) |
pdf |
| 18 |  | EWS:ERG-fusion(ETS)/CADO_ES1-EWS:ERG-ChIP-Seq(SRA014231)/Homer | 1e-41 | -9.636e+01 | 0.0000 | 2829.0 | 10.81% | 2165.1 | 8.38% | motif file (matrix) |
pdf |
| 19 |  | Ets1-distal(ETS)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-41 | -9.580e+01 | 0.0000 | 1219.0 | 4.66% | 800.6 | 3.10% | motif file (matrix) |
pdf |
| 20 |  | ETS(ETS)/Promoter/Homer | 1e-41 | -9.514e+01 | 0.0000 | 2018.0 | 7.71% | 1466.5 | 5.67% | motif file (matrix) |
pdf |
| 21 |  | ZNF711(Zf)/SHSY5Y-ZNF711-ChIP-Seq(GSE20673)/Homer | 1e-40 | -9.397e+01 | 0.0000 | 10506.0 | 40.13% | 9332.9 | 36.11% | motif file (matrix) |
pdf |
| 22 |  | AP-2gamma(AP2)/MCF7-TFAP2C-ChIP-Seq(GSE21234)/Homer | 1e-40 | -9.288e+01 | 0.0000 | 7048.0 | 26.92% | 6039.2 | 23.37% | motif file (matrix) |
pdf |
| 23 |  | SCL(bHLH)/HPC7-Scl-ChIP-Seq(GSE13511)/Homer | 1e-39 | -9.178e+01 | 0.0000 | 15125.0 | 57.78% | 13877.0 | 53.69% | motif file (matrix) |
pdf |
| 24 |  | Bach1(bZIP)/K562-Bach1-ChIP-Seq(GSE31477)/Homer | 1e-38 | -8.940e+01 | 0.0000 | 232.0 | 0.89% | 84.5 | 0.33% | motif file (matrix) |
pdf |
| 25 |  | EWS:FLI1-fusion(ETS)/SK_N_MC-EWS:FLI1-ChIP-Seq(SRA014231)/Homer | 1e-38 | -8.912e+01 | 0.0000 | 2621.0 | 10.01% | 2005.8 | 7.76% | motif file (matrix) |
pdf |
| 26 |  | SpiB(ETS)/OCILY3-SPIB-ChIP-Seq(GSE56857)/Homer | 1e-37 | -8.588e+01 | 0.0000 | 1157.0 | 4.42% | 769.2 | 2.98% | motif file (matrix) |
pdf |
| 27 |  | EBF1(EBF)/Near-E2A-ChIP-Seq(GSE21512)/Homer | 1e-36 | -8.487e+01 | 0.0000 | 5772.0 | 22.05% | 4884.3 | 18.90% | motif file (matrix) |
pdf |
| 28 |  | PU.1(ETS)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-36 | -8.379e+01 | 0.0000 | 2131.0 | 8.14% | 1594.1 | 6.17% | motif file (matrix) |
pdf |
| 29 |  | YY1(Zf)/Promoter/Homer | 1e-34 | -7.974e+01 | 0.0000 | 569.0 | 2.17% | 320.8 | 1.24% | motif file (matrix) |
pdf |
| 30 |  | SPDEF(ETS)/VCaP-SPDEF-ChIP-Seq(SRA014231)/Homer | 1e-32 | -7.571e+01 | 0.0000 | 4094.0 | 15.64% | 3377.8 | 13.07% | motif file (matrix) |
pdf |
| 31 |  | IRF4(IRF)/GM12878-IRF4-ChIP-Seq(GSE32465)/Homer | 1e-30 | -7.114e+01 | 0.0000 | 1745.0 | 6.67% | 1297.3 | 5.02% | motif file (matrix) |
pdf |
| 32 |  | Sox10(HMG)/SciaticNerve-Sox3-ChIP-Seq(GSE35132)/Homer | 1e-28 | -6.637e+01 | 0.0000 | 5386.0 | 20.57% | 4617.5 | 17.87% | motif file (matrix) |
pdf |
| 33 |  | Sox3(HMG)/NPC-Sox3-ChIP-Seq(GSE33059)/Homer | 1e-28 | -6.597e+01 | 0.0000 | 5846.0 | 22.33% | 5050.3 | 19.54% | motif file (matrix) |
pdf |
| 34 |  | AP-2alpha(AP2)/Hela-AP2alpha-ChIP-Seq(GSE31477)/Homer | 1e-28 | -6.508e+01 | 0.0000 | 5555.0 | 21.22% | 4782.2 | 18.50% | motif file (matrix) |
pdf |
| 35 |  | Foxo1(Forkhead)/RAW-Foxo1-ChIP-Seq(Fan et al.)/Homer | 1e-27 | -6.336e+01 | 0.0000 | 6357.0 | 24.28% | 5546.1 | 21.46% | motif file (matrix) |
pdf |
| 36 |  | bZIP:IRF(bZIP,IRF)/Th17-BatF-ChIP-Seq(GSE39756)/Homer | 1e-27 | -6.228e+01 | 0.0000 | 1693.0 | 6.47% | 1278.4 | 4.95% | motif file (matrix) |
pdf |
| 37 |  | NF-E2(bZIP)/K562-NFE2-ChIP-Seq(GSE31477)/Homer | 1e-23 | -5.399e+01 | 0.0000 | 238.0 | 0.91% | 113.4 | 0.44% | motif file (matrix) |
pdf |
| 38 |  | NFkB-p65(RHD)/GM12787-p65-ChIP-Seq(GSE19485)/Homer | 1e-23 | -5.365e+01 | 0.0000 | 2277.0 | 8.70% | 1823.7 | 7.06% | motif file (matrix) |
pdf |
| 39 |  | Sox4(HMG)/proB-Sox4-ChIP-Seq(GSE50066)/Homer | 1e-22 | -5.238e+01 | 0.0000 | 3011.0 | 11.50% | 2492.5 | 9.64% | motif file (matrix) |
pdf |
| 40 |  | Fosl2(bZIP)/3T3L1-Fosl2-ChIP-Seq(GSE56872)/Homer | 1e-21 | -5.035e+01 | 0.0000 | 980.0 | 3.74% | 701.5 | 2.71% | motif file (matrix) |
pdf |
| 41 |  | PU.1-IRF(ETS:IRF)/Bcell-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-21 | -4.997e+01 | 0.0000 | 5577.0 | 21.30% | 4891.3 | 18.93% | motif file (matrix) |
pdf |
| 42 |  | RUNX1(Runt)/Jurkat-RUNX1-ChIP-Seq(GSE29180)/Homer | 1e-20 | -4.829e+01 | 0.0000 | 3367.0 | 12.86% | 2838.1 | 10.98% | motif file (matrix) |
pdf |
| 43 |  | EBF(EBF)/proBcell-EBF-ChIP-Seq(GSE21978)/Homer | 1e-19 | -4.445e+01 | 0.0000 | 1157.0 | 4.42% | 868.5 | 3.36% | motif file (matrix) |
pdf |
| 44 |  | Bach2(bZIP)/OCILy7-Bach2-ChIP-Seq(GSE44420)/Homer | 1e-18 | -4.261e+01 | 0.0000 | 637.0 | 2.43% | 434.4 | 1.68% | motif file (matrix) |
pdf |
| 45 |  | Smad3(MAD)/NPC-Smad3-ChIP-Seq(GSE36673)/Homer | 1e-17 | -4.061e+01 | 0.0000 | 9611.0 | 36.71% | 8828.4 | 34.16% | motif file (matrix) |
pdf |
| 46 |  | FOXP1(Forkhead)/H9-FOXP1-ChIP-Seq(GSE31006)/Homer | 1e-17 | -3.968e+01 | 0.0000 | 1314.0 | 5.02% | 1020.5 | 3.95% | motif file (matrix) |
pdf |
| 47 |  | AR-halfsite(NR)/LNCaP-AR-ChIP-Seq(GSE27824)/Homer | 1e-16 | -3.906e+01 | 0.0000 | 12423.0 | 47.46% | 11589.4 | 44.84% | motif file (matrix) |
pdf |
| 48 |  | Jun-AP1(bZIP)/K562-cJun-ChIP-Seq(GSE31477)/Homer | 1e-16 | -3.727e+01 | 0.0000 | 728.0 | 2.78% | 522.2 | 2.02% | motif file (matrix) |
pdf |
| 49 |  | RUNX2(Runt)/PCa-RUNX2-ChIP-Seq(GSE33889)/Homer | 1e-16 | -3.708e+01 | 0.0000 | 2567.0 | 9.81% | 2160.2 | 8.36% | motif file (matrix) |
pdf |
| 50 |  | RUNX-AML(Runt)/CD4+-PolII-ChIP-Seq(Barski et al.)/Homer | 1e-15 | -3.665e+01 | 0.0000 | 2344.0 | 8.95% | 1958.9 | 7.58% | motif file (matrix) |
pdf |
| 51 |  | Stat3(Stat)/mES-Stat3-ChIP-Seq(GSE11431)/Homer | 1e-15 | -3.608e+01 | 0.0000 | 2115.0 | 8.08% | 1752.5 | 6.78% | motif file (matrix) |
pdf |
| 52 |  | p53(p53)/Saos-p53-ChIP-Seq(GSE15780)/Homer | 1e-15 | -3.497e+01 | 0.0000 | 325.0 | 1.24% | 199.0 | 0.77% | motif file (matrix) |
pdf |
| 53 |  | p53(p53)/Saos-p53-ChIP-Seq/Homer | 1e-15 | -3.497e+01 | 0.0000 | 325.0 | 1.24% | 199.0 | 0.77% | motif file (matrix) |
pdf |
| 54 |  | Nrf2(bZIP)/Lymphoblast-Nrf2-ChIP-Seq(GSE37589)/Homer | 1e-14 | -3.362e+01 | 0.0000 | 194.0 | 0.74% | 103.4 | 0.40% | motif file (matrix) |
pdf |
| 55 |  | RUNX(Runt)/HPC7-Runx1-ChIP-Seq(GSE22178)/Homer | 1e-14 | -3.348e+01 | 0.0000 | 2285.0 | 8.73% | 1920.9 | 7.43% | motif file (matrix) |
pdf |
| 56 |  | Fox:Ebox(Forkhead,bHLH)/Panc1-Foxa2-ChIP-Seq(GSE47459)/Homer | 1e-13 | -3.058e+01 | 0.0000 | 2734.0 | 10.44% | 2350.7 | 9.10% | motif file (matrix) |
pdf |
| 57 |  | MafA(bZIP)/Islet-MafA-ChIP-Seq(GSE30298)/Homer | 1e-13 | -3.041e+01 | 0.0000 | 2585.0 | 9.87% | 2213.7 | 8.57% | motif file (matrix) |
pdf |
| 58 |  | Sox2(HMG)/mES-Sox2-ChIP-Seq(GSE11431)/Homer | 1e-12 | -2.992e+01 | 0.0000 | 2975.0 | 11.36% | 2578.7 | 9.98% | motif file (matrix) |
pdf |
| 59 |  | Rbpj1(?)/Panc1-Rbpj1-ChIP-Seq(GSE47459)/Homer | 1e-12 | -2.929e+01 | 0.0000 | 5330.0 | 20.36% | 4805.1 | 18.59% | motif file (matrix) |
pdf |
| 60 |  | PRDM1(Zf)/Hela-PRDM1-ChIP-Seq(GSE31477)/Homer | 1e-12 | -2.908e+01 | 0.0000 | 2192.0 | 8.37% | 1858.2 | 7.19% | motif file (matrix) |
pdf |
| 61 |  | NFkB-p65-Rel(RHD)/ThioMac-LPS-Expression(GSE23622)/Homer | 1e-12 | -2.906e+01 | 0.0000 | 265.0 | 1.01% | 163.0 | 0.63% | motif file (matrix) |
pdf |
| 62 |  | AP-1(bZIP)/ThioMac-PU.1-ChIP-Seq(GSE21512)/Homer | 1e-12 | -2.897e+01 | 0.0000 | 2040.0 | 7.79% | 1719.3 | 6.65% | motif file (matrix) |
pdf |
| 63 |  | Ap4(bHLH)/AML-Tfap4-ChIP-Seq(GSE45738)/Homer | 1e-12 | -2.893e+01 | 0.0000 | 4613.0 | 17.62% | 4126.5 | 15.97% | motif file (matrix) |
pdf |
| 64 |  | STAT4(Stat)/CD4-Stat4-ChIP-Seq(GSE22104)/Homer | 1e-12 | -2.841e+01 | 0.0000 | 3292.0 | 12.58% | 2884.5 | 11.16% | motif file (matrix) |
pdf |
| 65 |  | Sox6(HMG)/Myotubes-Sox6-ChIP-Seq(GSE32627)/Homer | 1e-12 | -2.770e+01 | 0.0000 | 5016.0 | 19.16% | 4519.2 | 17.49% | motif file (matrix) |
pdf |
| 66 |  | Bcl6(Zf)/Liver-Bcl6-ChIP-Seq(GSE31578)/Homer | 1e-11 | -2.755e+01 | 0.0000 | 4672.0 | 17.85% | 4193.4 | 16.23% | motif file (matrix) |
pdf |
| 67 |  | STAT6(Stat)/Macrophage-Stat6-ChIP-Seq(GSE38377)/Homer | 1e-11 | -2.542e+01 | 0.0000 | 1751.0 | 6.69% | 1473.4 | 5.70% | motif file (matrix) |
pdf |
| 68 |  | Fra1(bZIP)/BT549-Fra1-ChIP-Seq(GSE46166)/Homer | 1e-10 | -2.480e+01 | 0.0000 | 1558.0 | 5.95% | 1300.5 | 5.03% | motif file (matrix) |
pdf |
| 69 |  | ETS:RUNX(ETS,Runt)/Jurkat-RUNX1-ChIP-Seq(GSE17954)/Homer | 1e-10 | -2.392e+01 | 0.0000 | 445.0 | 1.70% | 318.3 | 1.23% | motif file (matrix) |
pdf |
| 70 |  | RXR(NR),DR1/3T3L1-RXR-ChIP-Seq(GSE13511)/Homer | 1e-10 | -2.315e+01 | 0.0000 | 4492.0 | 17.16% | 4059.4 | 15.71% | motif file (matrix) |
pdf |
| 71 |  | Stat3+il21(Stat)/CD4-Stat3-ChIP-Seq(GSE19198)/Homer | 1e-9 | -2.158e+01 | 0.0000 | 2558.0 | 9.77% | 2244.2 | 8.68% | motif file (matrix) |
pdf |
| 72 |  | Tbx20(T-box)/Heart-Tbx20-ChIP-Seq(GSE29636)/Homer | 1e-9 | -2.157e+01 | 0.0000 | 686.0 | 2.62% | 532.6 | 2.06% | motif file (matrix) |
pdf |
| 73 |  | STAT6(Stat)/CD4-Stat6-ChIP-Seq(GSE22104)/Homer | 1e-9 | -2.143e+01 | 0.0000 | 1726.0 | 6.59% | 1472.0 | 5.70% | motif file (matrix) |
pdf |
| 74 |  | PR(NR)/T47D-PR-ChIP-Seq(GSE31130)/Homer | 1e-9 | -2.120e+01 | 0.0000 | 6662.0 | 25.45% | 6160.6 | 23.84% | motif file (matrix) |
pdf |
| 75 |  | Atf3(bZIP)/GBM-ATF3-ChIP-Seq(GSE33912)/Homer | 1e-9 | -2.112e+01 | 0.0000 | 1827.0 | 6.98% | 1567.9 | 6.07% | motif file (matrix) |
pdf |
| 76 |  | CEBP:AP1(bZIP)/ThioMac-CEBPb-ChIP-Seq(GSE21512)/Homer | 1e-9 | -2.079e+01 | 0.0000 | 2174.0 | 8.30% | 1892.0 | 7.32% | motif file (matrix) |
pdf |
| 77 |  | GRHL2(CP2)/HBE-GRHL2-ChIP-Seq(GSE46194)/Homer | 1e-8 | -2.054e+01 | 0.0000 | 1080.0 | 4.13% | 887.8 | 3.44% | motif file (matrix) |
pdf |
| 78 |  | Tcfcp2l1(CP2)/mES-Tcfcp2l1-ChIP-Seq(GSE11431)/Homer | 1e-8 | -1.963e+01 | 0.0000 | 557.0 | 2.13% | 426.5 | 1.65% | motif file (matrix) |
pdf |
| 79 |  | Smad2(MAD)/ES-SMAD2-ChIP-Seq(GSE29422)/Homer | 1e-8 | -1.877e+01 | 0.0000 | 5891.0 | 22.50% | 5443.5 | 21.06% | motif file (matrix) |
pdf |
| 80 |  | Tcf12(bHLH)/GM12878-Tcf12-ChIP-Seq(GSE32465)/Homer | 1e-8 | -1.860e+01 | 0.0000 | 3992.0 | 15.25% | 3624.4 | 14.02% | motif file (matrix) |
pdf |
| 81 |  | MyoD(bHLH)/Myotube-MyoD-ChIP-Seq(GSE21614)/Homer | 1e-8 | -1.856e+01 | 0.0000 | 3198.0 | 12.22% | 2870.3 | 11.11% | motif file (matrix) |
pdf |
| 82 |  | Chop(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer | 1e-7 | -1.831e+01 | 0.0000 | 676.0 | 2.58% | 535.5 | 2.07% | motif file (matrix) |
pdf |
| 83 |  | Atoh1(bHLH)/Cerebellum-Atoh1-ChIP-Seq(GSE22111)/Homer | 1e-7 | -1.811e+01 | 0.0000 | 3964.0 | 15.14% | 3602.2 | 13.94% | motif file (matrix) |
pdf |
| 84 |  | CHR(?)/Hela-CellCycle-Expression/Homer | 1e-7 | -1.809e+01 | 0.0000 | 2046.0 | 7.82% | 1790.4 | 6.93% | motif file (matrix) |
pdf |
| 85 |  | TATA-Box(TBP)/Promoter/Homer | 1e-7 | -1.807e+01 | 0.0000 | 4123.0 | 15.75% | 3755.0 | 14.53% | motif file (matrix) |
pdf |
| 86 |  | PPARE(NR),DR1/3T3L1-Pparg-ChIP-Seq(GSE13511)/Homer | 1e-7 | -1.738e+01 | 0.0000 | 3582.0 | 13.68% | 3245.3 | 12.56% | motif file (matrix) |
pdf |
| 87 |  | BATF(bZIP)/Th17-BATF-ChIP-Seq(GSE39756)/Homer | 1e-7 | -1.644e+01 | 0.0000 | 1756.0 | 6.71% | 1531.9 | 5.93% | motif file (matrix) |
pdf |
| 88 |  | n-Myc(bHLH)/mES-nMyc-ChIP-Seq(GSE11431)/Homer | 1e-6 | -1.560e+01 | 0.0000 | 2503.0 | 9.56% | 2238.3 | 8.66% | motif file (matrix) |
pdf |
| 89 |  | p53(p53)/mES-cMyc-ChIP-Seq(GSE11431)/Homer | 1e-6 | -1.547e+01 | 0.0000 | 118.0 | 0.45% | 70.3 | 0.27% | motif file (matrix) |
pdf |
| 90 |  | T1ISRE(IRF)/ThioMac-Ifnb-Expression/Homer | 1e-6 | -1.501e+01 | 0.0000 | 65.0 | 0.25% | 32.2 | 0.12% | motif file (matrix) |
pdf |
| 91 |  | Myf5(bHLH)/GM-Myf5-ChIP-Seq(GSE24852)/Homer | 1e-6 | -1.458e+01 | 0.0000 | 2571.0 | 9.82% | 2311.3 | 8.94% | motif file (matrix) |
pdf |
| 92 |  | Olig2(bHLH)/Neuron-Olig2-ChIP-Seq(GSE30882)/Homer | 1e-6 | -1.455e+01 | 0.0000 | 6104.0 | 23.32% | 5700.0 | 22.05% | motif file (matrix) |
pdf |
| 93 |  | NFkB-p50,p52(RHD)/Monocyte-p50-ChIP-Chip(Schreiber et al.)/Homer | 1e-6 | -1.413e+01 | 0.0000 | 770.0 | 2.94% | 637.7 | 2.47% | motif file (matrix) |
pdf |
| 94 |  | Foxa2(Forkhead)/Liver-Foxa2-ChIP-Seq(GSE25694)/Homer | 1e-6 | -1.382e+01 | 0.0000 | 2201.0 | 8.41% | 1968.2 | 7.62% | motif file (matrix) |
pdf |
| 95 |  | NFAT(RHD)/Jurkat-NFATC1-ChIP-Seq(Jolma et al.)/Homer | 1e-5 | -1.354e+01 | 0.0000 | 2795.0 | 10.68% | 2533.0 | 9.80% | motif file (matrix) |
pdf |
| 96 |  | STAT1(Stat)/HelaS3-STAT1-ChIP-Seq(GSE12782)/Homer | 1e-5 | -1.332e+01 | 0.0000 | 1037.0 | 3.96% | 885.8 | 3.43% | motif file (matrix) |
pdf |
| 97 |  | Smad4(MAD)/ESC-SMAD4-ChIP-Seq(GSE29422)/Homer | 1e-5 | -1.291e+01 | 0.0000 | 5910.0 | 22.58% | 5533.4 | 21.41% | motif file (matrix) |
pdf |
| 98 |  | E2F4(E2F)/K562-E2F4-ChIP-Seq(GSE31477)/Homer | 1e-5 | -1.288e+01 | 0.0000 | 3274.0 | 12.51% | 2996.1 | 11.59% | motif file (matrix) |
pdf |
| 99 |  | Gata4(Zf)/Heart-Gata4-ChIP-Seq(GSE35151)/Homer | 1e-5 | -1.268e+01 | 0.0000 | 2803.0 | 10.71% | 2549.0 | 9.86% | motif file (matrix) |
pdf |
| 100 |  | Max(bHLH)/K562-Max-ChIP-Seq(GSE31477)/Homer | 1e-5 | -1.256e+01 | 0.0000 | 2091.0 | 7.99% | 1875.6 | 7.26% | motif file (matrix) |
pdf |
| 101 |  | MafK(bZIP)/C2C12-MafK-ChIP-Seq(GSE36030)/Homer | 1e-5 | -1.175e+01 | 0.0000 | 703.0 | 2.69% | 588.5 | 2.28% | motif file (matrix) |
pdf |
| 102 |  | Gata2(Zf)/K562-GATA2-ChIP-Seq(GSE18829)/Homer | 1e-4 | -1.140e+01 | 0.0000 | 1756.0 | 6.71% | 1569.5 | 6.07% | motif file (matrix) |
pdf |
| 103 |  | Gata1(Zf)/K562-GATA1-ChIP-Seq(GSE18829)/Homer | 1e-4 | -1.113e+01 | 0.0000 | 1573.0 | 6.01% | 1400.0 | 5.42% | motif file (matrix) |
pdf |
| 104 |  | Atf4(bZIP)/MEF-Atf4-ChIP-Seq(GSE35681)/Homer | 1e-4 | -1.099e+01 | 0.0000 | 821.0 | 3.14% | 700.7 | 2.71% | motif file (matrix) |
pdf |
| 105 |  | p63(p53)/Keratinocyte-p63-ChIP-Seq(GSE17611)/Homer | 1e-4 | -1.057e+01 | 0.0001 | 1111.0 | 4.24% | 971.3 | 3.76% | motif file (matrix) |
pdf |
| 106 |  | CArG(MADS)/PUER-Srf-ChIP-Seq(Sullivan et al.)/Homer | 1e-4 | -1.022e+01 | 0.0001 | 894.0 | 3.42% | 772.6 | 2.99% | motif file (matrix) |
pdf |
| 107 |  | STAT5(Stat)/mCD4+-Stat5-ChIP-Seq(GSE12346)/Homer | 1e-4 | -9.954e+00 | 0.0001 | 1162.0 | 4.44% | 1023.1 | 3.96% | motif file (matrix) |
pdf |
| 108 |  | MITF(bHLH)/MastCells-MITF-ChIP-Seq(GSE48085)/Homer | 1e-4 | -9.927e+00 | 0.0001 | 2952.0 | 11.28% | 2721.8 | 10.53% | motif file (matrix) |
pdf |
| 109 |  | NF1-halfsite(CTF)/LNCaP-NF1-ChIP-Seq(Unpublished)/Homer | 1e-3 | -9.082e+00 | 0.0003 | 6257.0 | 23.90% | 5928.5 | 22.94% | motif file (matrix) |
pdf |
| 110 |  | RBPJ:Ebox(?,bHLH)/Panc1-Rbpj1-ChIP-Seq(GSE47459)/Homer | 1e-3 | -8.574e+00 | 0.0005 | 1173.0 | 4.48% | 1044.6 | 4.04% | motif file (matrix) |
pdf |
| 111 |  | c-Myc(bHLH)/mES-cMyc-ChIP-Seq(GSE11431)/Homer | 1e-3 | -8.572e+00 | 0.0005 | 1655.0 | 6.32% | 1499.4 | 5.80% | motif file (matrix) |
pdf |
| 112 | 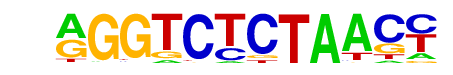 | PRDM14(Zf)/H1-PRDM14-ChIP-Seq(GSE22767)/Homer | 1e-3 | -8.143e+00 | 0.0007 | 956.0 | 3.65% | 844.9 | 3.27% | motif file (matrix) |
pdf |
| 113 |  | Oct4:Sox17(POU,Homeobox,HMG)/F9-Sox17-ChIP-Seq(GSE44553)/Homer | 1e-3 | -7.977e+00 | 0.0008 | 366.0 | 1.40% | 301.4 | 1.17% | motif file (matrix) |
pdf |
| 114 |  | Pit1(Homeobox)/GCrat-Pit1-ChIP-Seq(GSE58009)/Homer | 1e-3 | -7.909e+00 | 0.0009 | 2483.0 | 9.49% | 2296.4 | 8.89% | motif file (matrix) |
pdf |
| 115 |  | Isl1(Homeobox)/Neuron-Isl1-ChIP-Seq(GSE31456)/Homer | 1e-3 | -7.825e+00 | 0.0009 | 5551.0 | 21.20% | 5263.7 | 20.37% | motif file (matrix) |
pdf |
| 116 |  | Ascl1(bHLH)/NeuralTubes-Ascl1-ChIP-Seq(GSE55840)/Homer | 1e-3 | -7.503e+00 | 0.0013 | 6350.0 | 24.26% | 6047.1 | 23.40% | motif file (matrix) |
pdf |
| 117 |  | MyoG(bHLH)/C2C12-MyoG-ChIP-Seq(GSE36024)/Homer | 1e-3 | -7.104e+00 | 0.0019 | 4062.0 | 15.52% | 3830.5 | 14.82% | motif file (matrix) |
pdf |
| 118 |  | Tcf3(HMG)/mES-Tcf3-ChIP-Seq(GSE11724)/Homer | 1e-3 | -7.077e+00 | 0.0019 | 684.0 | 2.61% | 599.0 | 2.32% | motif file (matrix) |
pdf |
| 119 |  | ETS:E-box(ETS,bHLH)/HPC7-Scl-ChIP-Seq(GSE22178)/Homer | 1e-3 | -6.932e+00 | 0.0022 | 323.0 | 1.23% | 267.5 | 1.04% | motif file (matrix) |
pdf |
| 120 |  | RORgt(NR)/EL4-RORgt.Flag-ChIP-Seq(GSE56019)/Homer | 1e-2 | -6.545e+00 | 0.0032 | 354.0 | 1.35% | 297.3 | 1.15% | motif file (matrix) |
pdf |
| 121 |  | c-Myc(bHLH)/LNCAP-cMyc-ChIP-Seq(Unpublished)/Homer | 1e-2 | -6.533e+00 | 0.0032 | 2007.0 | 7.67% | 1857.1 | 7.19% | motif file (matrix) |
pdf |
| 122 |  | NFAT:AP1(RHD,bZIP)/Jurkat-NFATC1-ChIP-Seq(Jolma et al.)/Homer | 1e-2 | -6.371e+00 | 0.0037 | 500.0 | 1.91% | 432.8 | 1.67% | motif file (matrix) |
pdf |
| 123 |  | Nr5a2(NR)/mES-Nr5a2-ChIP-Seq(GSE19019)/Homer | 1e-2 | -6.245e+00 | 0.0042 | 1808.0 | 6.91% | 1670.4 | 6.46% | motif file (matrix) |
pdf |
| 124 |  | HNF4a(NR),DR1/HepG2-HNF4a-ChIP-Seq(GSE25021)/Homer | 1e-2 | -5.964e+00 | 0.0055 | 1433.0 | 5.47% | 1315.3 | 5.09% | motif file (matrix) |
pdf |
| 125 |  | NeuroD1(bHLH)/Islet-NeuroD1-ChIP-Seq(GSE30298)/Homer | 1e-2 | -5.591e+00 | 0.0079 | 2791.0 | 10.66% | 2625.5 | 10.16% | motif file (matrix) |
pdf |
| 126 |  | CEBP:CEBP(bZIP)/MEF-Chop-ChIP-Seq(GSE35681)/Homer | 1e-2 | -5.502e+00 | 0.0085 | 467.0 | 1.78% | 407.2 | 1.58% | motif file (matrix) |
pdf |
| 127 |  | USF1(bHLH)/GM12878-Usf1-ChIP-Seq(GSE32465)/Homer | 1e-2 | -5.355e+00 | 0.0098 | 1550.0 | 5.92% | 1434.2 | 5.55% | motif file (matrix) |
pdf |
| 128 |  | GFY-Staf(?,Zf)/Promoter/Homer | 1e-2 | -5.217e+00 | 0.0112 | 250.0 | 0.96% | 209.2 | 0.81% | motif file (matrix) |
pdf |
| 129 |  | Unknown-ESC-element(?)/mES-Nanog-ChIP-Seq(GSE11724)/Homer | 1e-2 | -5.110e+00 | 0.0123 | 2343.0 | 8.95% | 2200.3 | 8.51% | motif file (matrix) |
pdf |
| 130 |  | Oct2(POU,Homeobox)/Bcell-Oct2-ChIP-Seq(GSE21512)/Homer | 1e-2 | -5.015e+00 | 0.0135 | 805.0 | 3.08% | 728.2 | 2.82% | motif file (matrix) |
pdf |
| 131 |  | CTCF-SatelliteElement(Zf?)/CD4+-CTCF-ChIP-Seq(Barski et al.)/Homer | 1e-2 | -4.972e+00 | 0.0140 | 44.0 | 0.17% | 29.6 | 0.11% | motif file (matrix) |
pdf |
| 132 |  | PAX3:FKHR-fusion(Paired,Homeobox)/Rh4-PAX3:FKHR-ChIP-Seq(GSE19063)/Homer | 1e-2 | -4.624e+00 | 0.0196 | 500.0 | 1.91% | 444.9 | 1.72% | motif file (matrix) |
pdf |







































































































































































































































































