
More Information | Similar Motifs Found

More Information | Similar Motifs Found

More Information | Similar Motifs Found
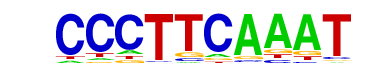
More Information | Similar Motifs Found

More Information | Similar Motifs Found

More Information | Similar Motifs Found

More Information | Similar Motifs Found

More Information | Similar Motifs Found

More Information | Similar Motifs Found

More Information | Similar Motifs Found

More Information | Similar Motifs Found

More Information | Similar Motifs Found

More Information | Similar Motifs Found

More Information | Similar Motifs Found

More Information | Similar Motifs Found
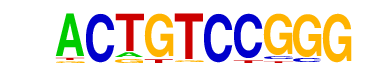
More Information | Similar Motifs Found

More Information | Similar Motifs Found

More Information | Similar Motifs Found

More Information | Similar Motifs Found

More Information | Similar Motifs Found

More Information | Similar Motifs Found